The Taxonomic Analysis and Genetic Biodiversity of Nodule Bacteria Soybean (Glycine Max (L.) Merr.) Isolated from Soils of Central Asia
ABSTRACT
The taxonomy of nitrogen-fixing bacteria forming symbiotic associations with legumes has undergone profound changes in recent years. The use of very sensitive and precise molecular methods has made it possible to identify a wide variety of rhizobia, especially among the natural field populations of these soil bacteria. The purpose of the present the study was aimed at identifying and characterizing isolated soybean rhizobia from different types of soils of Central Asia that are regularly used for agricultural production. The actual composition and genetic diversity of the population of natural fields has been studied using various PCR methods such as 16S rDNA. Seventy rhizobia isolates were characterized and compared with reference and / or type strains representing Bradyrhizobium japonicum, Bradyrhizobium elkanii and Sinorhizobium fredii, Pantoea agglomerans, Enterobacter cloacae emerging in α - β- γ- protobacteria.
KEYWORDS
Nitrogen fixation; Rhizobia; Soybean; Indigenous strains; Bradyrhizobium japonicum; DNA fingerprinting; Genetic diversity
INTRODUCTION
Rhizobia are usually defined as nitrogen-fixing soil bacteria capable of inducing the formation of root or stem nodules on leguminous plants in which atmospheric nitrogen is reduced to ammonia for the benefit of the plant. Due to their considerable agricultural and environmental significance, these legume symbionts have been extensively studied. During the last years, the assessment of diversity within rhizobial natural populations in various regions of the world has received increased attention to determine the actual composition and characteristics of indigenous strains isolated from different cultivated legumes [1-4]. The development of numerous molecular genetic methods has greatly contributed to these investigations. The availability of several sensitive and accurate PCR-based genotyping methods [5,6] has enabled the differentiation among closely related bacterial strains and the detection of a higher rhizobial diversity than previously considered [7,8].
Symbiotic nitrogen fixing bacteria are represented by a phylogenetically disparate class of alpha- and beta-proteobacteria - usually collectively termed rhizobia - that have achieved the function of fixing atmospheric nitrogen (N2) in symbiosis with legumes. The majority of the symbiotic species are represented in the alpha-proteobacteria order Rhizobiales, which, amongst many others, contain the agriculturally important nitrogen fixing genera of Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium and Azorhizobium. One impediment to the broader use of rhizobia in agriculture is the production of compatible inoculants. There are substantial host, strain and environmental specificities that limit the use of potentially important legume fodder and crops as alternatives to nitrogen fertilizers [9]. To date, very few rhizobial strains have been fully sequenced, accounting for less than 1% of all bacterial genomes available today. In this study, the genetic biodiversity of the soybean nodule bacteria was examined. Various instruments have been evaluated in comparison, and the results may also contribute to further research in the future large-scale comparative genomic studies. The main aim of the present study was to identify and characterise the indigenous soybean rhizobia isolated from the most important soybean growing areas in Central Asia, and to assess genetic diversity of natural field population by using different PCR-based methods.
MATERIALS AND METHODS
DNA Preparation
Isolation and purification of nodule bacteria from soybean plants was written in our previous work [10]. Bacterial DNA was obtained by the method of J.Marmur with some modification [11]. The nucleotide sequence of the PCR product and Bioinformatics methods RDP were carried out at Ben-Gurion University (Israel).
PCR Amplification of the 16S rRNA Gene
The 16S rRNA gene from nodule bacteria of Soya bean was amplified using universal primers 1070f (59-ACGGGCGGTGTGTAC-39) and 1392r(59-CGCCCGCCGCGCCC CGCGCCCGGCCCGCCGCCCCCGCCCCACGGGCGGTGTGTAC39) [12]. Each PCR mixture contained the following: 10pmol each primer, 200μM dNTPs, 1U Tag DNA polymerase, 100-200ng genomic DNA and Taq polymerase buffer in a final reaction volume of 50μl. The DNA thermal cycler used for PCR amplification was programmed as follows: an initial extensive denaturation step at 94 °Cfor 5 min; 30 cycles of 94°C for 1 min, 53°C for 1 min and 72°C for 1.5 min; and a final extension step at 72°C for 10 min.
Phylogenetic Analysis
The complete 300-354-bp 16S rRNA gene sequences were compared with the sequences available in the GenBank database using the standard Basic Local Alignment Search Tool, BLASTn [13], at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). From the aligned sequences, neighbor joining dendrograms were constructed with the software Ribosomal Database Project (RDP-II) [14]. and software MEGA version 4.0.2. [15]. The robustness of the inferred trees was evaluated by 1000 bootstrap resampling’s.
RESULTS
The RDP-II obtains rRNA sequence data from the major International Nucleotide Sequence Databases (GenBank/EMBL/ DDBJ). These sequences are provided aligned and annotated format. Release 8.1 contains 16 277 archaeal and bacterial, 1503 mitochondrial, and 5201 eukaryotic small subunit rRNA sequences. In order to provide a phylogenetic context for the data, RDP-II makes available over 100 trees that span the phylogenetic breadth of life. Most of these trees created using the WEIGHBOR algorithm, with submitted alignments.
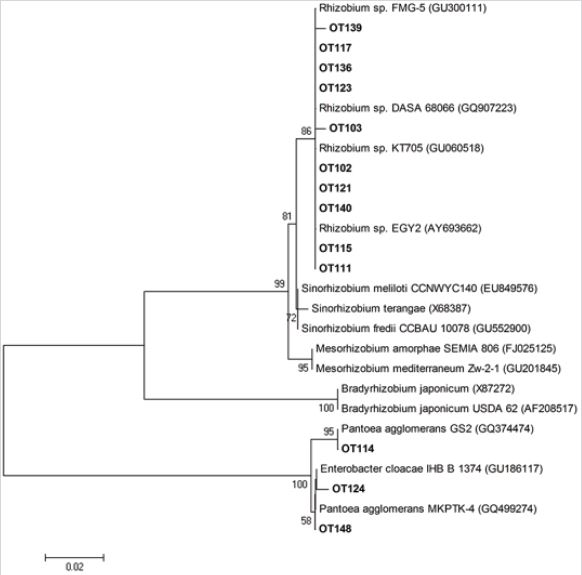
In our studies, the isolates were numbered from T-1 (OT) to T-70, OC1-OC-50 and all of them were used in our studies. Taxonomic analysis of bacterial isolates of nodule bacteria isolated from nodules of soybean plants was studied using the 16S rRNA gene method. Determination of the nucleotide sequence of the 16S rRNA gene of nodule bacteria in soybeans makes it possible to reveal the specificity of bacteria to the genus, and some bacteria to the species. The results of a comparative BLAST analysis of the nucleotide conserved region of the 16S rRNA gene of bacteria from soybeans were 99% identical to the genes of Rhizobium sp. EGY2 (AY693662.1) Interestingly, the nucleotide indicates isolates T-2. T-3. T-11, T-17 and T-18 from soybeans by 96-98% to the genes of Sinorhizobium meliloti CCNWY49C. (AB535707.1), Sinorhizobium fredii CCBAU 10078 (GU552900.1), Mesorhizobium mediterraneum Zw-2-1 (GU201845.1), Bacteria Mesorhizobium obiense Zw-1 (GU201844.1), Bradyrhizobium japonicum PRY65 (AF239848.2). The conserved region of the 16S rRNA gene of bacteria T-5, T-9, and T-12 coincided by 98-99% with the genes of Pantoea agglomerans GS2 (GQ374474.1), Enterobacter cloacae IHB B 1374 (GU186117.1) and bacteria Pantoea agglomerans MKPTK-4 (GQ499274.1), respectively. Similar results were obtained for the nodule bacteria from the root nodules of two soybean (Glycine max) cultivars [16]. Analysis of the 16S rRNA gene of nodule bacteria Soy shows that many nucleotide isolates show 98-99% coincidence with the genes of the Rhizobium phaseoli R620 (CPO13542.1) strain. Moreover, the 16S rRNA genes of the isolates were 97- 99% homologous to the genes of several bacterial species, such as Sinorhizbium meliloti CCNWY C140 (EU849576.1), Sinorhizobium meliloti YcS2 (AB535707.1), Sinorhizobium (GU), 552900.1 Bacteria Mesorhizobium tianshanense (FM203306.1), Mesorhizobium amorphae CCNWYC131 (EU849577.1) and Bradyrhizobium japonicum PRY62 (AF 239847.2).
The OC112 bacterium is 99% identical to the nucleotide sequences of the 16S rRNA gene of Burkholderia caryophylli WAB1944 (AM184283.1), the genes from T-36 to T51 coincide by 97% with the genes of Pantoea agglomerans HXJ (HM016799.1), and by 98%. coincided with the genes of Enterobacter sp. RF-100 (GQ205104.1). When analyzing the phylogenetic tree created on the basis of the nucleotide sequence of the studied 16S rRNA gene of various bacteria, it was found that these bacteria from soybeans were related to the class Alphaproteobacteria. and some isolates belonged to the class Gammaproteobacteria (Figure 1-3). The bacteria included in the 1st cluster were Alphaproteobacteria, the 2nd cluster - Betaproteobacteria, the 3rd cluster were Gammaproteobacteria, in particular to the Genera Enterobacter and Panoea.
CONCLUSION
Rhizobia soybeans isolated from different sections of the field in Central Asia have been characterized using DNA-basic methods. PCR analysis showed the taxonomy of the 16S rDNA region of the genome defining them to species, and undoubtedly showed that all field isolates differed from Bradyrhizobium japonicum and especially from Sinorhizobium fredii type of strains. Although most soy isolates were closely related to the Bradyrhizobium japonicum strain, seven isolates made up a separate group of strains. Our results show that this divergent group should be considered as Bradyrhizobium japonicum strains. However, to confirm this statement and further genetic studies are needed to clarify the status of these isolates. In future publications, we will present more detailed data on the genetic biodiversity of soybean nodule bacteria isolated from different soils of Central Asia. Our results overlap with other scientists’ findings on nodule bacteria that have shown that there are other types of nitrogen-fixing in the soyabean nodule bacteria and our data only supplement such data [16].
REFERENCES
- Black M, Moolhuijzen P, Chapman B, Barrero R, Howieson J, et al. (2012) The genetics of symbiotic nitrogen fixation: Comparative genomics of 14 rhizobia strains by resolution of protein clusters. Genes 3(1): 138-166.
- Gresshoff PM, Ferguson BJ (2017) Molecular signals in nodulation control. Int J Mol Sci 18(1): 10-12.
- Tewari S, Arora NK (2016) 2- Soybean production under flooding stress and its mitigation using plant growth-promoting microbes. Environ Stresses Soybean Prod 2: 23-40.
- Jaspers E, Overmann,J (2014) Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol 70(8): 4831-4839.
- Koskey G, Mburu SW, Kimiti JM, Ombori O, Maingi JM et al. (2018) Genetic characterization and diversity of rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.) varieties. Front Microbiol 9: 968.
- Li YH, Wang R, Zhang XX, Young JPW, Wang ET, et al. (2015) Bradyrhizobium guangdongense sp. nov. and Bradyrhizobium guangxiense sp. nov., isolated from effective nodules of peanut. Int J Syst Evol Microbiol 65(12): 4655-4661.
- Li YH, Wang R., Sui XH, Wang ET, Zhang XX, et al. (2019) Bradyrhizobium nanningense sp. nov., Bradyrhizobium guangzhouense sp. nov. and Bradyrhizobium zhanjiangense sp. nov., isolated from effective nodules of peanut in Southeast China. Syst Appl Microbiol 42: 126002.
- Cardoso AA, Andraus MP, Borba TC, Martin-Didonet CC, Ferreira EP (2017) Characterization of rhizobia isolates obtained from nodules of wild genotypes of common bean. Braz J Microbiol 48(1): 43-50.
- Liao J, Liu X, Hu A, Song H, Chen X (2020) Effects of biochar‑based controlled release nitrogen fertilizer on nitrogen use efficiency of oilseed rape (Brassica napus L.). Scientific Reports 10: 11063.
- Umarov B (2021) New rhizobial bacteria of the genus Sinorhizobium fredii entering symbiosis with soybean plants. Open Acc J Biomedical Sci 3(6):942-944.
- Aronshtam A, Umarov B, Erko V, Andronov E, Simarov B (1993) Use of a Rhizobium meliloti cosmid gene bank for cloning the leucine biosynthesis gene, which is involved in regulating the development of nitrogen-fixing symbiosis with alfalfa Soviet genetics. 29(2): 185-192.
- Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62(2): 340-346.
- Blast. Command line applications user manual. National Center for Biotechnology. Bethesda, Maryland, USA.
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, et al. (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Research 31(1): 442-443.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24(8): 1596-1599.
- Habibi S, Ayubi A, Ohkama-Ohtsu N, Sekimoto H, Yokoyama TI (2017) Genetic characterization of soybean rhizobia isolated from different ecological zones in north-eastern Afghanistan. Microbes Environ 32(1): 71-79.
Article Type
Research Article
Publication history
Received date: September 13, 2021
Published date: October 04, 2021
Address for correspondence
Umarov BR, Tashkent Research Institute of Vaccines and Serum, Ministry of Health of the Republic of Uzbekistan
Copyright
©2021 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Umarov BR, Boymatova M. The Taxonomic Analysis and Genetic Biodiversity of Nodule Bacteria Soybean (Glycine Max (L.) Merr.) Isolated from Soils of Central Asia. 2021- 3(5) OAJBS.ID.000326.
Figure 1: The phylogenetic tree constructed using nucleotide sequences of 16sRNA (300-350bp) from of 20 isolates according to the RDP- II program.
Figure 2: The phylogenetic tree constructed using nucleotide sequences of 16sRNA (300-350bp) from of 20 isolates according to the RDP-II program.
Figure 3: Phylogenetic tree, 16S rRNA gene of soybean nodule bacterial isolates. Alphaproteobacteria; b- Betaproteobacteria.