Absence of Mammarenavirus RNA among their Natural Rodent and Potential other Reservoirs in Wildlife in Gabon
ABSTRACT
LCMV RNA has been detected both in its natural host reservoir, the house mouse Mus musculus, and other rodent species in Gabon. In addition, many pathogenic and nonpathogenic Mammarenavirus reservoirs are also present in the country; suggesting that other arenaviruses should be present together with new virus hosts. Considering the importance of rodent-born arenaviruses for human health, we look for the presence of these viruses in rodents, bats and bushmeats collected in the entire Gabonese territory. Here we failed to detect arenaviruses RNA in all the tested samples. This absence can be explained by several hypotheses: (i) the geographical hypothesis, (ii) the genetic structure of the host that can impact viral distribution, (iii) the relationship between host density and the presence of the virus, (iv) the dilution effect, and (v) the detection method used. To get more information about the circulation of arenaviruses and their host reservoirs in Gabon, it is necessary to both enrich collected animals and make additional analyses. The enrichment is related to the increased number and diversity of animal species and the extension of the collection to all periods of the year. Additional analyses should be: (i) the use of complementary specific multiplex systems of PCR targeting both segments of arenaviruses, (ii) serological analyses and Whole Genome Sequencing technologies.
KEYWORDS
Arenavirus; Potential reservoirs; Rodents; Bats; Bushmeats; Gabon
ABBREVIATIONS
LASV: Lassa Virus; LCMV: Lymphocytic Choriomeningitis Virus; CNER: National Ethics Committee for Research; CIRMF: Centre Interdisciplinaire de Recherches Médicales de Franceville; DNA: Desoxyribonucleic Acid; RNA: Ribonucleic Acid
INTRODUCTION
Viruses of the family Arenaviridae are enveloped viruses with a genome consisting of two single-stranded RNA segments, designated small (S) and large (L). The family is divided into three genera: Mammarenavirus which have rodents and bats as natural reservoirs; Hartmanivirus and Reptarenavirus [1] which have snakes as natural reservoirs. Some viruses from the Mammarenavirus genus led to severe hemorrhagic fever outbreaks in Sub-Saharan Africa (Lassa virus (LASV) and Lujo virus) and in Central and South America (Junin virus, Machupo virus, Sabia virus, Charape virus and Guanarito virus). Particularly, Lujo virus (an emerging arenavirus found in South Africa) killed 4 of the 5 people who were infected [2] and Lassa hemorrhagic fever is responsible each year for thousands of cases and deaths in several countries of West Africa, including mainly Nigeria, Sierra Leone, Liberia and Guinea, where it is endemic [3]. In Central Africa, antibodies against LASV have been found in rodents in Cameroon [4], and in humans in Congo [5], two neighboring countries of Gabon. In addition, a recent study conducted in Gabon found antibodies against another arenavirus, the LCMV, among the human population [6].
The principal host reservoirs of LASV are the multimammate rats Mastomys natalensis and Mastomys erythroleucus. Preliminary studies shown that Mastomys natalensis is present in Gabon (personal data). Other pathogenic and non-pathogenic arenaviruses reservoir rodents have also been found in the country including Mus musculus (the natural host reservoir of Lymphocytic Choriomeningitis Virus (LCMV [7])), Praomys sp (the reservoir of Mobala and Ippy viruses), Lemniscomys striatus, Malacomys sp (other known hosts of Mobala and Ippy viruses [8]), Mus Nannomys sp (reservoir of Lunk, Gbagroube and kodoko viruses [9-11]), Hylomyscus sp (the reservoir of the Menekre virus [11]), R. rattus and R. norvegicus (found infected with the Wenzhou virus in China [12]) [6,13-15] and personal data. Recently, LCMV ARN has been identified both in its natural reservoir, the house mouse Mus musculus domesticus, a native species from Asia which would have been newly introduced into the country together with the virus [15], and others native rodents’ species [6]. Considering the risk factor of arenavirus infection to humans, the detection of LCMV antibodies in the Gabonese population, and the presence of natural arenavirus reservoirs in Gabon, combined with the recent discovery of LCMV in a new host (Atherurus africanus) [6], there is an urgent need to explore the circulation of these viruses among their natural reservoirs and investigate new arenavirus reservoirs to assess the potential of the circulation of LASV or other arenaviruses to wildlife in Gabon. With the ultimate objective of avoiding the infection of humans with these viruses.
MATERIALS AND METHODS
This project was carried out with the agreement of the National Ethics Committee for Research (CNER) of Gabon (N°0020/CNE/ SG/P). The samples were done by a veterinary team and coordinated by a veterinary expert FELASA (No.2011-38/VetAgroSup).
Animal Collection
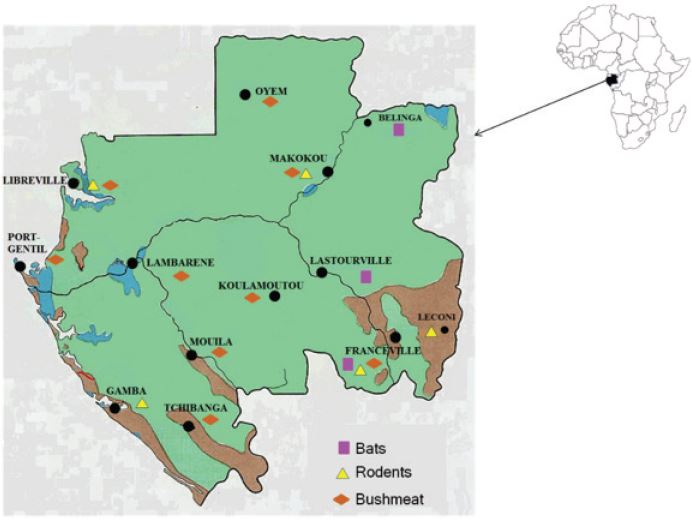
Samples were collected throughout the country (Figure 1). Bats and rodents were caught in three and four provinces respectively whereas bushmeat samples were collected throughout the country near the nine biggest cities. Rodents were caught as previously described using a standard live-trapping protocol [13,14]. Bats capture was made with nets installed at the cave entrances just before twilight as previously described [16,17]. For bushmeat, animals were bought in special bushmeat markets, where legally hunted bushmeat is usually sold. For all samples from small and big mammals, the capture, sampling, and exportation were made with the permission of local authorities.
Species Identification
The identification of bats and bushmeat species was made by trained field biologists. For rodents, morphological identification was made using identification keys [13,18,19] while molecular identification was done using a partial sequence of mitochondrial cytochrome b gene [15,20] and microsatellites for Rattus rodents previously used for a population genetic study [14].
Captured bats and rodents were removed carefully as soon as possible to minimize injury, drowning, strangulation, or stress according to the guidelines of the American Society of Mammalogists [21]. Safe euthanasia was practiced according to the Felasa protocol and autopsy was performed. Samples of brain, feces, heart, intestine, kidney, liver, lung, and spleen were collected, frozen individually in the cryotubes and transported to the CIRMF (Centre Interdisciplinaire de Recherches Médicales de Franceville) laboratory where they were stored at -80 °C until molecular analyses.
RNA Extraction from Organs
Livers and spleens were pooled and crushed in 600μl of cold phosphate buffered saline (Ambion) for 2min at 1500 strakes/min in a ball-mill tissue grinder (2000 Geno/grinder, SpexCertiprep). One hundred microliters of this suspension were incubated in 300μl of Qiagen lysis buffer for 10min and total RNA was extracted from this mixture using EZ1 RNA Tissue mini kit and the EZ1 BioRobot automat both from Qiagen, in the presence of DNase (Qiagen) and according to the manufacturer’s instructions.
Virus Molecular Detection
Two conventional One Step RT-PCR systems without nested step have been used for the search of all arenaviruses RNA. Analyses were performed on total RNA extracted from five pooled samples from the same species of rodents, bats and bushmeats. If pooled samples were positive, these samples were then tested individually.
The first conventional PCR system used generic primers ARS16V (5’-GGCATWGANCCAAACTGATT-3’) and ARS1 (5’-CGCACCGGGGATCCTAGGC-3’) targeting the glycoprotein gene of arenavirus with a modified protocol from Lozano et al. [22]. Reverse-transcription was performed separately using High- Capacity cDNA Reverse Transcription kit from Applied Biosystems according to the manufacturer’s instructions; followed by PCR using SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen by life technologies) in the final reaction volume of 25μl, containing 5μl of cDNA, 12.5μl of 2x Reaction Mix, 0.4μM of each primer and 1μl of SuperScript III RT/ Platinum TaqMix. Samples were heated to 95 °C for 2min, followed by 40 cycles of 30s at 95 °C, 30s at 55 °C and 1min at 72 °C, with a final extension step at 72 °C for 5min.
The second conventional one step RT-PCR system used generic primers LVL3754A-minus (5’-CACATCATTGGTCCCCATTTACTATGATC) and LVL3359Aplus (5’-AGAATTAGTGAAAGGGAGAGCAATTC) targeting the RNA polymerase of arenaviruses with a modified protocol from Lecompte et al. [9]. Reverse transcription was also performed using SuperScript III (Invitrogen by life technologies) followed by PCR in a final volume of 25μl containing 5μl of RNA, 12.5μl of 2x Reaction Mix, 0.4μM of each primer and 1μl of SuperScript III RT/Platinum TaqMix with the same condition that the first conventional PCR system used. Cycling conditions were the following: 45min at 50 °C, 94 °C for 2min, followed by 45 cycles of 30s at 94 °C, 1min at 60 °C and 2min at 68 °C, final extension step at 68 °C for 7min and an additional hold at 20 °C for 2min.
For all conventional PCR systems, samples at the expected size (640bp for the first system and about 480bp for the second system) after amplification, were purified (when necessary) directly from agarose gel using QIAquick Gel Extraction kit (Qiagen) and sent for sequencing at SeqLab (Göttigen, Germany).
RESULTS
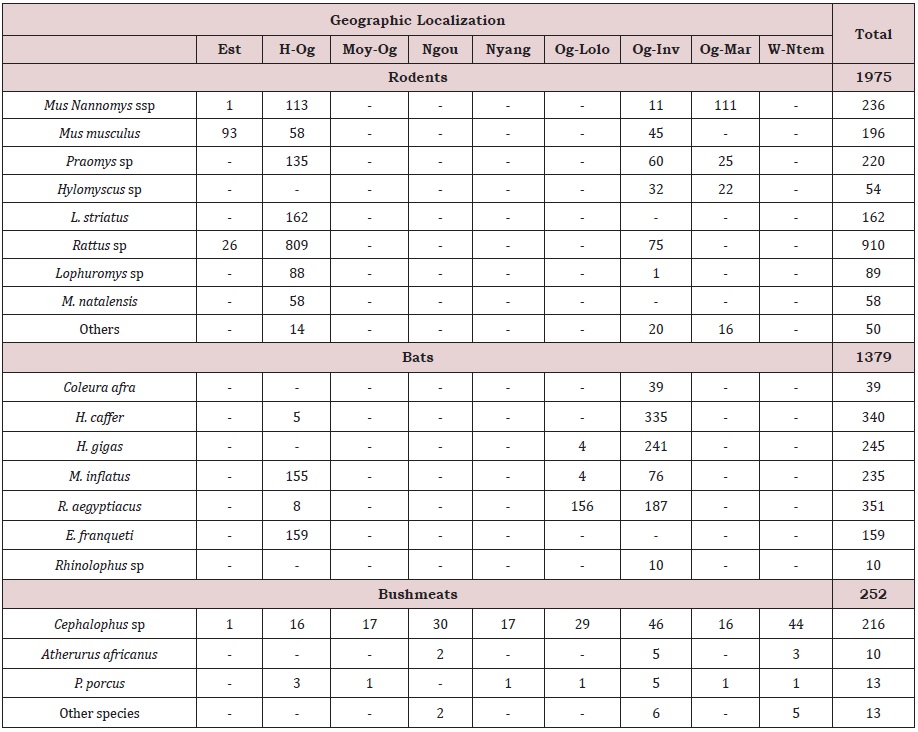
From 2009 up to 2014, livers and spleens were collected from a total of 1,379 bats, 252 bushmeats and 1,975 rodents and shrews. Collected animals were seven species from both bats and bushmeats, 17 genera among rodents and one shrew species (Table 1).
Among bats, Rousettus aegyptiacus (351 individuals) was the most collected species, followed by H. caffer (340 individuals), H. gigas (245 individuals) and M. inflattus (235 indviduals)). Cephalophus sp was the most frequently encountered species among bushmeats (Table 1). The most representative genera among rodents were imported species (Rattus sp (rattus and norvegicus) (910 individuals), and Mus musculus domesticus (196 individuals)) which all account for nearly 56% of all collected rodents and one native species (Mus Nannomys ssp (236 individuals)). Other collected native species include Praomys sp, Hylomyscus sp, M. natalensis, Lemniscomys striatus and Malacomys longipes (Table 1), all are known as natural reservoirs of arenaviruses. The majority of these native rodent species were found outside human dwelling both in semi-urban and rural areas whereas imported species were mostly caught inside human dwellings in urban (Libreville) and semi-urban areas (Franceville and Makokou) (Table 1).
We looked for arenavirus RNA in livers and spleens from all the 3606 collected animals by the two PCR systems. Unfortunately, all the analyses were negatives suggesting that none of the tested samples were infected by arenaviruses.
DISCUSSION
Although during this study we found both invasive and native rodent species, the latter ones are poorly represented and live far from human dwelling. That should be due to the introduction of invasive species which tend to push away from housing native species and dominate them when invading an area [23]. However, we cannot exclude that the trapping method (inside and around human dwelling only) could also result in this difference. So, for the future, it will be necessary to catch rodents not only in and around human dwellings, but also in forests and savannas in both semiurban and rural areas to get more specimens by species.
Negative PCR results obtained in bats are in line with all the studies performed on these animal species, strengthening the hypothesis that bats probably are not reservoirs for arenaviruses [24]. In fact, a recent study conducted in 2017 in Trinidad, found no RNA of arenaviruses in bats [25]. However, the detection of antibodies specific to the Tascaribe virus (TCRV) nucleocapsid antigen in Artibeus jamaicensis during that study [25] clearly showed that bats could be naturally exposed, even infected, with arenavirus. In addition, experimental infection of Jamaican Fruit Bat by TCRV caused substantial mortality and morbidity in bats [24]. These findings indicate that further studies need to be undertaken to elucidate the role of bats within the natural cycle of some arenaviruses. However, few individuals have been caught for some bat species and sampling was done only in three Gabonese provinces. Thus, the results we observed should be due to the size and restricted geographical location of samples. Thus, we need increase and diversify species sampling all around the country and add new species, such as insectivorous bats.
In view of the recent discovery of LCMV ARN in Atherurus africanus [6] and regarding arenaviruses discovery in new hosts such as snakes [26] and shrews [12], research was conducted on bushmeat during this study. Unfortunately, our results were negative. Nevertheless, we caught only 10 Atherurus africanus animals, 4 shrews and no snakes during this study. To our knowledge, in the literature, only one study detected LCMV RNA in only one Atherurus africanus animal among the 18 tested during that study [6] and no other animal species had a positive RNA or antibodies result; suggesting that the infection of these animals is rare. Our study and the previous one [6] constitutes preliminary studies in this field and need to be strengthen by a bigger number of animals sampled.
Rodents are natural reservoirs of the Arenaviridae family, so one would expect to find mammarenaviruses among the tested animals. However, we failed to detect any virus despite the presence of wellknown natural arenavirus reservoirs like M. natalensis, Praomys sp, M. Nannomys ssp, Lemniscomys striatus, Malacomys longipes, Hylomyscus sp, Rattus rattus, Rattus norvegicus and Mus musculus domesticus in collected animals. These results are in accordance with one previous Gabonese study [13] and could be explained by a number of hypotheses.
The first one is the geographical hypothesis, which postulates that viruses are sometimes present only in a restricted part of their host’s geographic range [27]. Some arenaviruses have not been detected in all geographical areas occupied by their reservoir even if for the majority of viruses, the geographical distribution matches with the distribution of his natural host [27,28]. Indeed, since its discovery in 1969, LASV is constrained to Sub-Saharan West Africa [29,30], despite the presence of his host throughout the African continent [31]. In the same way, Mopeia, Morogoro and Gairo viruses have been detected only in the eastern part of Africa [32- 34] and many other arenaviruses have been detected only in one region across the entire African continent [9-11,35-37]; suggesting that in the Arenaviridae family, many viruses should be restricted to one part of the African continent. This restriction if not only limited to some parts of the African continent but is also applicable to one country [27]. This could explain why previous studies found LCMV in different hosts in Gabon [6,15], while another one [13] and the present study failed to detect any viruses.
This geographic hypothesis would be explained by the second hypothesis: viral distribution can be impacted by the genetic structure of the host [27,38]. M. natalensis, one of the most widespread rodents in Sub-Saharan Africa, displays six divergent mitochondrial phylogroups differently distributed across the Africa: A-I phylogroup in Western part, A-II in the center, A-III, B-IV and B-V in the Eastern part and finally B-VI in the Southern part [27,39]. These different phylogroups also carry different Mammarenavirus species. LASV restricted to West Africa is carried by the M. natalensis’ phylogroup A-I and A-II (only from the west part of the Niger river for this last phylogroup) [40], suggesting a natural geographic barrier for virus dispersion [41,42]; a Mobalalike virus is carried by the phylogroup A-II [40] while other viruses restricted to the Eastern and Southern parts of Africa are carried by the B-IV (Gairo virus) [27,33,43], B-V (Morogoro virus) [27,34,43] and B-VI (both Mopeia [44] and Luna [27,45] viruses) phylogroups. The fact that some phylogroups (A-I, B-IV, B-V, and B-VI) virus reservoirs are not present in Gabon would explain the absence of viruses like Gairo, LASV, Luna and Morogoro.
The third hypothesis is that there is a relationship between host density and the presence of the virus; this means that a virus may be absent where the host density is below a persistence threshold [27,46]. A study made in Tanzania shown that among 1,772 rodents from the species M.natalensis tested for the arenavirus, only 52 were positive (2.93%) [27], confirming that the prevalence of mammarenaviruses among their natural reservoirs is very low. The size of our animal collection is small and for many of these animals, we collected less than 40 individuals by species. That would not be enough to detect positive animals. It should be necessary to increase the number of animals by species.
The fourth hypothesis is that the dilution effect, that is to say rodent richness, would have a protective effect on the geographical expansion of viruses [42,47], as previously demonstrated for LASV [42]. Rodent species richness showed a significant negative association with Lassa fever emergence [42]. A high diversity of rodent species could suppress the activities of reservoir rodents and reduce the likelihood of spillover transmission from rodents. High rodent species diversity could decrease the migration of infected rodents from the disease endemic area [42,47]. The dilution effect could occur when there are a great deal of host species available, such as in our study, as 17 rodent genera were caught.
In this study, we used degenerated primers which allow the detection of all known members of the Arenaviridae family, but also decrease the sensibility of detections. Thus, we need to use a multiplex system, which will allow for more specific detections.
Furthermore, serological information by screening antibodies such as immunoglobulin G could help with detecting previous infections, thus providing further insight on the circulation of arenaviruses in Gabon. Indeed, several previous studies support the acute infection model that postulates that small mammals’ infection by arenaviruses are normally acute, which means that (i) there is an increase of mortality among infected rodents [28,32,48,49], (ii) viral clearance operates quickly and, (iii) is replaced by antibodies production among survivals [50-52]. Thus, tested individuals are positive either by PCR or IgG [53].
CONCLUSION
Even if all molecular analyses for the detection of arenaviruses RNA in our sample collection were negative, we cannot exclude the circulation of other arenaviruses besides the LCMV in Gabon. Ultimately, in this study we caught many rodent species reservoirs of arenaviruses (Mastomys natalensis, Hylomyscus sp, Rattus sp, Mus musculus, Praomys sp, Lemniscomys striatus, Malacomys sp and Mus Nannomys) that were found infected in neighboring countries like Central African Republic. Therefore, to gain further insight from investigations of animal reservoirs of arenaviruses in Gabon, it will be necessary to add new technologies like antibodies detection and Next Generation Sequencing, together with the increase of the number of animals by species. Indeed, in faraway geographical areas from endemic areas, few reservoir species are found infected and/or the viral load is too low to be detectable by PCR.
ACKNOWLEDGEMENT
CIRMF is supported by the Government of Gabon, Total-Fina-Elf Gabon, and the Ministère des Affaires Etrangères et Européennes of France. We thank all the people involved in sample collection, especially André Délicat, Philippe Engandja and Ida Lepagna.
FUNDING
This work was supported by the Gabonese government.
AUTHOR’S CONTRIBUTION
NN and EML contributed to the design of the study; NN, JBMP, GDM, OLBME, HDS, LB and BN collected the samples, NN, JBMP, OLBME and HDS performed the laboratory analyses, NN and EML prepared the first draft of the manuscript, NN and EML supervised the work.
REFERENCES
- Maes P, Alkhovsky SV, Bào Y, Martin B, Monica B, et al. (2018) Taxonomy of the family Arenaviridae and the order Bunyavirales: Update 2018. Arch Virol 163(8): 2295-2310.
- Paweska JT, Sewlall NH, Ksiazek TG, Lucille HB, Martin JH, et al. (2009) Nosocomial outbreak of novel arenavirus infection, Southern Africa. Emerg Infect Dis 15(10): 1598-1602.
- Fichet CE, Rogers DJ (2009) Risk maps of lassa fever in West Africa. PLoS Negl Trop Dis 3(3): e388.
- Mylne AQN, Pigott DM, Longbottom J, Freya S, Kirsten AD, et al. (2015) Mapping the zoonotic niche of Lassa fever in Africa. Trans R Soc Trop Med Hyg 109(8): 483-492.
- Talani P, Konongo JD, Gromyko A, Nanga MJ, Yala F, et al. (1999) Prévalence des anticorps anti-fièvres hémorragiques d’origine virale dans la région du Pool (Congo-Brazzaville). Preval Viral Antibodies Haemorrh Fever Pool Area 46(8-9): 424-427.
- Ushijima Y, Abe H, Ozeki T, Georgelin NO, Marien JM, et al. (2021) Identification of potential novel hosts and the risk of infection with lymphocytic choriomeningitis virus in humans in Gabon, Central Africa. Int J Infect Dis 105: 452-459.
- Ledesma J, Fedele CG, Carro F, Lourdes L, María Paz S, et al. (2009) Independent lineage of lymphocytic choriomeningitis virus in wood mice (Apodemus sylvaticus), Spain. Emerg Infect Dis. 15(10): 1677-1680.
- Salazar-Bravo J, Ruedas L, Yates TL (2002) Mammalian reservoirs of arenaviruses. Curr Top Microbiol Immunol 262: 25-63.
- Lecompte E, ter Meulen J, Emonet S, Daffis S, Charrel RN (2007) Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus Nannomys minutoides) in West Africa. Virology 364(1): 178- 183.
- Akihiro I (2011) Novel arenavirus, Zambia. Emerging Infectious Disease Journal 17(10): 1921-1924.
- Coulibaly GD, Allali B, Kouassi SK, et al. (2011) Novel arenavirus sequences in hylomyscus sp. and Mus (Nannomys) setulosus from côte d’ivoire: Implications for evolution of arenaviruses in Africa. PLoS One 6(6): e20893.
- Li K, Lin XD, Wang W, Mang S, Wen PG, et al. (2015) Isolation and characterization of a novel arenavirus harbored by Rodents and Shrews in Zhejiang province, China. Virology 476: 37-42.
- Mangombi JB, N’dilimabaka N, Lekana-Douki JB, Octavie B, Sydney MN, et al. (2021) First investigation of pathogenic bacteria, protozoa and viruses in rodents and shrews in context of forest-savannah-urban areas interface in the city of Franceville (Gabon). PLoS One 16(3): 1-28.
- Mangombi JB, Brouat C, Loiseau A, Banga O, Leroy EM, et al. (2016) Urban population genetics of the invasive black rats in Franceville, Gabon. J Zool 299(3): 183-190.
- N′Dilimabaka N, Berthet N, Rougeron V, Joa BM, Patrick D, et al. (2015) Evidence of Lymphocytic Choriomeningitis Virus (LCMV) in Domestic Mice in Gabon: Risk of Emergence of LCMV Encephalitis in Central Africa. J Virol 89(2): 1456-1460.
- Maganga GD, Bourgarel M, Ebang Ella G, Jan FD, Jean PG, et al. (2011) Is marburg virus enzootic in Gabon? J Infect Dis 204(Suppl 3): 13-16.
- Drexler JF, Corman VM, Müller MA (2012) Bats host major mammalian paramyxoviruses. Nat Commun 3: 796.
- Duplantier J (1989) Les rongeurs myomorphes forestiers du Nord-Est du Gabon: Structure du peuplement, démographie, domaines vitaux. Rev d’écologie 44(4): 329-346.
- Nicolas V, Schaeffer B, Missoup AD, Jan K, Marc C, et al. (2012) Assessment of three mitochondrial genes (16S, Cytb, CO1) for identifying species in the Praomyini tribe (Rodentia: Muridae). PLoS One 7(5): 1-11.
- Maarit J, Jeremy BS (2002) Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol Ecol 11(12): 2613-2621.
- (1987) Animal care and use. American Society of Mammalogists.
- Lozano ME, Posik DM, Albariño CG, Schujman G, Ghiringhelli PD, et al. (1997) Characterization of arenaviruses using a family-specific primer set for RT-PCR amplification and RFLP analysis. Its potential use for detection of uncharacterized arenaviruses. Virus Res 49(1): 79-89.
- Ba K, Piry S, Ambroise D, Cédric Lippens, Christophe D, et al. (2015) Range expansion of the invasive house mouse Mus musculus domesticus in Senegal, West Africa : A synthesis of trapping data over three decades, 1983-2014. Mammal Review 45(3): 176-190.
- Cogswell-Hawkinson A, Bowen R, James S, David G, Charles HC, et al. (2012) Tacaribe virus causes fatal infection of an ostensible Reservoir host, the Jamaican fruit bat. J Virol 6(10): 5791-5799.
- Malmlov A, Seetahal J, Carrington C, Vernie R, Jerome F, et al. (2017) Serological evidence of arenavirus circulation among fruit bats in Trinidad. Published online 12(9): e0185308.
- Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, et al. (2012) Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: Candidate etiological agents for snake inclusion body disease. M Bio 3(4): 180-112.
- Cuypers LN, Baird SJE, Ha A (2020) Three arenaviruses in three subspecific natal multimammate mouse taxa in Tanzania : same host specificity, but different spatial genetic structure? 6(2): 1-12.
- Zapata JC, Salvato MS (2013) Arenavirus variations due to host-specific adaptation. Viruses 5(1): 241-278.
- Richmond JK, Baglole DJ (2003) Lassa fever: Epidemiology, clinical features, and social consequences. BMJ 327(7426): 1271-1275.
- Gibb R, Moses LM, Redding DW, Jones KE (2017) Understanding the cryptic nature of Lassa fever in West Africa. Pathog Glob Health 111(6): 276-288.
- Granjon L (2016) Mastomys natalensis (errata version published in 2017). IUCN Red List Threat Species 2016: e.T12868A1.
- Borremans B, Leirs H, Gryseels S, Günther S, Makundi R, et al. (2011) Presence of mopeia virus, an African arenavirus, related to biotope and individual rodent host characteristics: Implications for virus transmission. Vector-Borne Zoonotic Dis 11(8): 1125-1131.
- Gryseels S, Rieger T, Oestereich L, Bart C, Benny B, et al. (2015) Gairo virus, a novel arenavirus of the widespread Mastomys natalensis: Genetically divergent, but ecologically similar to Lassa and Morogoro viruses. Virology 476: 249-256.
- Günther S, Hoofd G, Charrel R, Röser C, Becker-Ziaja B, et al. (2009) Mopeia virus-related arenavirus in natal multimammate mice, Morogoro, Tanzania. Emerg Infect Dis 15(12): 2008-2012.
- Ishii A, Thomas Y, Moonga L, Nakamura I, Ohnuma A, et al. (2012) Molecular surveillance and phylogenetic analysis of Old World arenaviruses in Zambia. J Gen Virol 93(Pt 10): 2247-2251.
- Blasdell KR, Duong V, Eloit M, Fabrice C, Sowath L, et al. (2016) Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia. Elife 5: e13135.
- Witkowski PT, Kallies R, Hoveka J, Brita A, Ndapewa LI, et al. (2015) Novel arenavirus isolates from namaqua rock mice, Namibia, Southern Africa. Emerg Infect Dis 21(7): 1213-1216.
- Saxenhofer M, Schmidt S, Ulrich RG, Id GH (2019) Secondary contact between diverged host lineages entails ecological speciation in a European hantavirus. Published online 17(2): e3000142.
- Colangelo P, Verheyen E, Leirs H, Caroline T, Christiane D, et al. (2013) A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biological Journal of the Linnean Society 180(4): 901-916.
- Olayemi A, Obadare A, Oyeyiola A, Joseph I, Ayobami F, et al. (2016) Arenavirus diversity and phylogeography of Mastomys natalensis Rodents, Nigeria. 694-697.
- Siddle KJ, Eromon P, Barnes KG, Samar M, Judith UO, et al. (2018) Genomic analysis of lassa virus during an increase in cases in Nigeria in 2018. N Engl J Med 379(18): 1745-1753.
- Min KD, Hwang J, Schneider MC, So Y, Lee JY, et al. (2021) An exploration of the protective effect of rodent species richness on the geographical expansion of lassa fever in West Africa. PLoS Negl Trop Dis 15(2): 1-14.
- Gryseels S, Baird SJE, Borremans B, Makundi R, Leirs H (2017) When viruses don’t go viral: The importance of host phylogeographic structure in the spatial spread of arenaviruses. Published online 13(1):1-22.
- Wulff H, McIntosh BM, Hamner DB, Johnson KM (1977) Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ 55(4): 441-444.
- Biome LD (2010) Prevalence of a virus inducing behavioural manipulation near species range border. Published online 19(4): 2995- 3007.
- Ostfeld RS, Keesing F (2012) Effects of host diversity on infectious disease. Annu Rev Ecol Evol Syst 43: 157-182.
- Aires B, Aires B (1988) Is vertical transmission sufficient to maintain junin virus in nature? Gen Virol 69(6): 1437-1440.
- Alfredo DV, Vida LH, MSM (1987) Effect of persistent infection with junin virus on growth and reproduction of its natural reservoir, calomys musculinus. Am J Trop Med Hyg 37(3): 663-669.
- Fichet-calvet E, Becker-ziaja B (2014) Lassa serology in natural populations of rodents and horizontal transmission. Vector Borne Zoonotic Dis 14(9): 665-674.
- Borremans B, Gryseels S, Broecke BV, Joachim M, Rhodes M, et al. (2017) Arenavirus dynamics in experimentally and naturally infected rodents. Ecohealth 14(3): 463-473.
- Borremans B, Vossen R, Becker-Ziaja B (2015) Shedding dynamics of Morogoro virus, an African arenavirus closely related to Lassa virus, in its natural reservoir host Mastomys natalensis. Nat Publ Gr 1-8.
- Douglas KO, Cayol C, Forbes KM, Thelma AS, Olli V, et al. (2021) Serological evidence of multiple zoonotic viral infections among wild rodents in barbados. Pathogens 10(6): 663.
- Olayemi A, Oyeyiola A, Obadare A, Joseph I, Adetunji SA, et al. (2018) Widespread arenavirus occurrence and seroprevalence in small mammals, Nigeria. Parasit Vectors 11(1): 416.
Article Type
Research Article
Publication history
Received Date: February 21, 2022
Published: March 25, 2022
Address for correspondence
Nadine N’dilimabaka, Centre Interdisciplinaire de Recherches Médicales de Franceville (CIRMF), Département de Virologie, Unité Emergences des Maladies Virales, Gabon
Copyright
©2022 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Nadine N, Joa BMP, Gael DM, Octavie LBME, Huguette DS, etc. Absence of Mammarenavirus RNA among their Natural Rodent and Potential other Reservoirs in Wildlife in Gabon. 2022- 4(2) OAJBS.ID.000422.
Figure 1: Map of Gabon showing localizations where samples have been collected. The 9 provinces of Gabon were chosen because they present different ecosystems, a forest ecosystem (Koulamoutou, Lastoursville, Lembarené, Makokou and Oyem), a savanna ecosystem (Gamba, Léconi, Port-Gentil and Tchibanga,) both forest and savanna ecosystems (Bakoumba, Franceville and Mouila), and an urban environment (Libreville).
Table 1: Number and species distribution of sampled animals by localities in Gabon. For rodents and shrew, other species include Crocidura sp (4 individuals), Stochomys longicaudatus (5 individuals), Deomys ferrugineus (6 individuals), Hybomys univittatus (4 individuals), Cricetomys emini (11 individuals), Malacomys longipes (8 individuals), Heimyscus fumosus (8 individuals), Grammomys poensis (2 individuals) and Oenomys hypoxanthus (2 individuals); and for bushmeats other species comprise Nandinia binotata (3 individuals), Osteolaemus tetraspis (1 individual), Phataginus tricuspis (1 individual) and Manis gigantean (8 individuals).