Research Models in Biomedical Sciences: Advantages and Limitations
ABSTRACT
Research models are routinely used in the biomedical field when we are talking about assays. All these models have advantages and limitations and should be chosen carefully, to avoid, mainly, mistakes in ethical practices. However, it is a great challenge for researchers picking and classifying their experimental models due to the lack of clear literature definition or guidelines. Here, we pioneer describe the overall definition of the main research models and give a gamut of examples from the biomedical field. This review also provides recommendations for aligning experimental practices to meet the necessity of assessing an international research model classification.
KEYWORDS
Ex situ; Ex vivo; In silico; In situ; In vitro; In vivo
ABBREVIATIONS:
AIDS: Acquired Immunodeficiency Disease Syndrome; CAM: Chick Chorioallantoic Membrane; CCLE: Cancer Cell Line Encyclopedia; DNA: Deoxyribonucleic Acid; ELISA: Enzyme-Linked Immunosorbent Assay; FISH: Fluorescent In Situ Hybridization; HIV: Human Immunodeficiency Virus; iPCR: Immune Polymerase Chain Reaction; ISCT: In Silico Clinical Trial; MCL: Mantle Cell Lymphoma; OECD: Organization for Economic Co-operation and Development; PCOP: Porcine Cornea Opacity Permeability; PCR: Polymerase Chain Reaction; SPR: Surface Plasmon Resonance
INTRODUCTION
Latin remains a mainstay in academic literature, particularly in natural and medical sciences [1,2]. Indeed, a plethora of scientific expressions exist that are solely used in Latin [3], e.g. the experimental models in vitro, and in vitro [4,5], and in silico [6,7]. Whilst it is key to select the appropriate research model for a given hypothesis to avoid waste of time and resources [8], choosing a suitable model that is capable of meeting the study objectives, is often complicated by unclear or entirely lacking literature definition of the various models; often, further confusion is added due to semantic differences between languages [9]. This review provides an outline of the most used research models, i.e. in silico, in vitro, ex vivo, in situ, ex situ, and in vivo (Figure 1) and an in-depth discussion about the advantages and limitations of each model. Furthermore, examples of different technologies applied in biomedical sciences (e.g. drug discovery) surrounding the research models of sciences will be explored, aiming to adopt clear and unambiguous definitions within the employed models.
MAIN RESEARCH MODELS
In Silico
In silico models (i.e. performed on computer) always require digital technology to simulate a specific process (e.g. pharmacologic, physiologic, etc.). Originally, silico referred to the chemical element silicon (Si) or silicium, the main compound of computer circuits [10]. In fact, the scientific expression “in silico” was first described in 1989 for experiments carried out entirely on a computer [11]. Nevertheless, some authors still consider in silico studies as a virtual extension of in vivo and in vivo experiments [12,13]. In silico approaches are represented by techniques that use software to analyze data and often involve computational models or simulations based on existing information of closely related phenomena. The output can then be used to make predictions and suggest hypotheses as a basis for in vivo, ex vivo, and in vivo models [14].
The greatest advantage of in silico methods is that they are usually faster and cheaper than classical tests, whilst also reducing the number of animals to in vivo assays [15]. Nevertheless, there are several pitfalls that are commonly encountered when employing in silico assays. Mainly, in silico studies demand rigorous validation, including experimental verification during model development [16]. Nevertheless, researchers should not be concerned about using in silico approaches, which boost medical and pharmaceutical innovation, provide an affordable cost and allow huge business opportunities [17]. Thus, it is not surprising that the usage of in silico models has greatly increased in the last years, as demonstrated by pharmacokinetic, oncologic and drug discovery studies [18,19]. Indeed, following the development of ever improving bioinformatic tools, there was a greatly increase in the arsenal of in silico methodologies, which have already resulted in significantly improved prediction and elucidation of the dynamics surrounding complex biological phenomena [20,21].
Molecular docking, which explores protein-protein or protein-small molecule binding, is one of the preferred in silico methodologies used, especially in drug discovery [22]. Since its discovery in the early 1980s [23], more than 60 different docking tools and programs have been developed for both academy and companies [24]. Other in silico methods that are routinely used in research laboratories include molecular modelling (a technique used to model or mimic the structure of molecules) and protein sequencing and its alignment (methods used to evaluate identities and similarities in the amino acid sequence of proteins) [25-28].
In silico toxicity prediction is also being employed to great success [29]. The computational methods employed to predict the toxicity profile of drugs has stood out to design new medicines once as in vitro and in vivo methods are often limited by ethics committees, time-consuming, and require a lot of resources [30]. So far, more than 15 sources of in silico toxicity model data are available, including ToxCast, Tox21, and ToxBank [30]. Similarly, in silico bioprospecting is being explored for searching new molecules for different targets and applications [31]. Detailed in silico methods to design biotherapeutics (e.g. antibodies and vaccines) have been discussed in detail elsewhere [32].
Recently, the in silico clinical trial (ISCT) came to the light. This approach refers to the development of patient-specific models to form virtual cohorts for testing the safety and/or efficacy of new drugs and of new medical devices. Clinical trials involving new drugs are frequently classified into four phases (I-IV). Phase I recruits about 20 to 100 volunteers and aims to test safety and dose scalation. The use of ISCT can predict the optimal dosage. In Phase II trials, which recruit about 100 to 300 volunteers, ISCT could be used to find representative virtual patients that show adverse effects [33]. In the most expensive Phase III, conducted with 300 to 3,000 patients, ISCT can assess the efficacy in terms of reduced side effects, predict ineffectiveness, and reduce the number of enrolled patients. Thus, ISCT can meaningfully lower the costs related to Phase III clinical trial [33,34].
In Vitro
In vitro (i.e. within the glass) refers to a model in which the assay occurs outside of a living organism and in a controlled environment. In addition, in vitro techniques are very valuable for ethical (does not require animals) and economic reasons [35]. Although the number of in vitro assays could be uncountable, few examples frequently used will be explored in this section.
All analytical techniques are classified as in vitro techniques such as chromatographic and spectrometric techniques, although they can be used to evaluate samples from ex vivo and in vivo studies. Chromatography is a century-old technique [36] used to separate and quantify complex mixtures of molecules according to their properties, which can be classified in different types (e.g. liquid chromatography, gas chromatography, ionexchange chromatography, and affinity chromatography) [37- 39]. Moreover, chromatography can be regularly associated with mass spectrometry (MS) analyzes, which is a powerful analytical technique for identifying, quantifying, and exploring molecular structures [40]. Both techniques can be employed to different compounds, including volatile components, lipids, organic molecules, and proteins [41,42]. Another method used to separate proteins according to their Physico-chemical characteristics (charge and size) is electrophoresis [43,44]. Electrophoresis can be performed in a gel [45] or on a filter paper [46], besides been used combined with mass spectrometry to identify protein sequence and mass (i.e. in-gel digestion) [47].
There are also other important in vitro methods broadly used for studying structure and biochemical characteristics of molecules, such as circular dichroism [48], protein quantification (e.g. Bradford and Lowry methods) [49,50], crystallography [51], as well as enzymatic assays, both colorimetric and turbidimetric, such as assays employed for phosphodiesterase [52,53] and hyaluronidase enzymes [54,55], respectively.
Other groundbreaking in vitro analytical test exhaustively used for research purposes, diagnostics, pharmacokinetic studies (i.e. drug monitoring) and for evaluating quality control of products is the enzyme-linked immunosorbent assay (ELISA), which employs enzyme-mediated color change and respective absorbance to quantify the presence of antigen-antibody mechanism [56]. ELISA method is globally employed because it is cheap, simple, specific, and quick to execute [56]. In connection with antigen-antibody binding, surface plasmon resonance (SPR) is also used to evaluate affinity of immunocomplexes [57].
Most of molecular biology assays are based on in vitro experiments, including the prominent polymerase chain reaction (PCR), which allows amplification of specific DNA segments through repetitive cycles of denaturation, hybridization, and polymerase extension [58,59]. In the recent years, the immune-PCR (iPCR) has frequently been employed. The iPCR method provides classical PCR sensitivity to objects traditionally detected by ELISA [60]. Phage display, an elegant in vitro technology in which a bacteriophage (i.e. a bacteria and archaea infecting virus) is used to evolve peptides and antibodies has been considered a robust technology for the discovery of therapeutic human antibodies against several diseases (e.g. autoimmune diseases and cancer) [59,61-63]. Still in molecular biology subject, microarray is an in vitro renowned technique applied for characterizing protein-protein, protein-nucleic acid, protein-small molecule, and antibody-antigen interactions. The technique has been used in both basic research (e.g. omics analysis) [64] and clinical applications (e.g. tumor markers) [65,66].
Although experiments conducted with living cells removed from humans or experimental animals should be considered an ex vivo method, cultures of lineage cells are better defined as in vitro approaches. Since 1912, when Carrel and his coworkers developed the first cell line from chicken embryo heart, cell lines have been extensively explored. However, their popularity increased in 1951, after the discovery of the immortalized HeLa cells, obtained from an adenocarcinoma of a patient named Henrietta Lacks [67]. Recently, cell lines, in special human cell lines, are generally used as a source of cells for high-throughput in vitro screening assays [68]. For instance, 1457 cancer human cell lines have been explored to predict drug sensitivity, which was defined as a Cancer Cell Line Encyclopedia (CCLE) [69,70]. Nevertheless, the actual number of generated cell lines is not reported and hard to estimate [71].
Similarly, there are other not obvious assays employing cells and organs described as in vitro approaches. The in vitro chicken enucleated eye test can detect the degree of eye irritation of certain compounds within six hours [72], which is considered an alternative to the in vivo Draize eye test method performed in rabbits [73]. Recently, an interesting new concept of in vitro assay emerged, named organs-in-a-chip, which describes a cell culturebased model system in which cells of different kinds are placed on small structures (chips). These chip-cells combinations can mimic the role of an organ or tissue, creating the organ-like structures. The organ-like system can be used for different applications such as drug toxicity screening [74], and it is expected that the technique could revolutionize early clinical trials in human patients by clinical-trials-on-chips [75].
Nevertheless, there is a continuous conflict between in vitro and ex vivo models [76]. Indeed, some authors use in vitro to refer to experiments performed within live cells (not cell lines) outside the organism, which should be best classified as an ex vivo approach. In any event, in vitro models undoubtedly provided an inexpensive and high-throughput alternative to in vivo research strategies. However, although in vitro models are fruitfully used in biological fields, extrapolation of the observed effects to animal models and humans can be considered weak and may lead to a misinterpretation of the data [77,78]. Indeed, researches that intend to use in vitro tests as an alternative to in vivo tests should have their approaches validated through scientific studies to assess the reliability and relevance of the method for the given purpose [79].
Ex Vivo
An ex vivo model designates a methodology defined by experiments with living cells or tissues taken from an organism and placed in an external environment under controlled conditions [80- 82]. The advantage of ex vivo experiments is that they are certainly useful to substitute the usage of whole animals as subjects, creating a high output research environment [80,81,83,84]. There are a gamut of studies in the literature employing ex vivo assays such as: ex vivo human gene therapy [85]; ex vivo cell cultivation such as within natural killers [86] and hematopoietic stem cells [87]; and analysis of organs ex vivo such as kidneys [88], spleen [89], heart, and lung [90].
Controversially, some authors still recognize an ex vivo method as a combination of the in vivo and in vitro models [78]. Indeed, due to the ambiguity with the terminology, researchers can easily be confused when classifying an ex vivo model. To clarify this issue, the well-known ex vivo gene therapy is a prime example. The ex vivo gene therapy is performed with cells isolated from a living organism, which are expanded/differentiated out of the living (with or without genetic modification) and further administered them back to the same or other living organism [91]. Although this technique clearly involves both in vivo and in vitro concepts, the best definition for this model should be ex vivo.
Other eminent ex vivo method is testing ocular corrosives on bovine corneas from slaughtered animals, a test recommended by the Organization for Economic Co-operation and Development (OECD) guideline [92]. An advantage of this assay is its speed, with results usually obtained within 24 hours [93]. The porcine cornea opacity permeability (PCOP) assay is other widely used approach in the ophthalmic research field, which can be considered better in comparison to bovine corneas, since the porcine cornea better resembles the human cornea [94]. The mouse retina is also an excellent ex vivo model for determining autophagic cell function [95]. Also, the use of the same material can provide oxygen consumption information, showing the mitochondrial reserve capacity of photoreceptors [96].
Although in vitro and ex vivo methods can be considered in many cases a potential scientific tool to study different biological effects, sometimes the obtained results can indicate inconsistencies with in vivo models, indicating a potential misleading nature of in vitro and ex vivo procedures [97]. One example of in vitro and ex vivo experiment could be seen in Figure 2.
In Situ and Ex Situ
In situ models (i.e. on site or in position) can also been stated by researchers as in loco (i.e. in place), with both referring to studies performed inside its original place or “on site”. In situ models are relevant in studying or identifying the active components and mechanisms in their place of origin, such as organs or tissues [98,99]. In medical research, this model is often used to evaluate genes and their respective translated proteins, such as a cytogenetic technique named fluorescent in situ hybridization (FISH), in which the genetic material can be assessed without being removed from the cell [100]. Basically, the FISH method uses fluorescently labelled probes that can bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity [101]. Therefore, FISH is considered a gold standard method for detection of gene alterations, a very useful tool for cancer diagnosis [102-104]. In situ neoplasia or carcinoma in situ is, by definition, the primal form of cancer and is characterized by being limited to the compartment corresponding to the cell of origin [105]. Notably, studies within in situ neoplasia have allowed the elucidation of tumor formation mechanisms [98,106,107]. In fact, in lymphomas, premalignant stages have been identified by evaluating in situ follicular lymphoma (FL) and mantle cell lymphoma (MCL) [107]. Several other studies regarding in situ carcinoma can be found elsewhere [106,108,109].
However, in situ models cannot be applied to all cell types, for example lymphocytes, which exhibit a circulating nature and cannot be studied in loco [98]. In the pharmacokinetics field, in situ models have also been explored for drug absorption tests [110- 112]. In drug absorption evaluation through the intestinal barrier, the in situ intestinal model showed to be even more accurate than the ex vivo model [112,113]. This technique consists of a cannula introduction into the small intestine of anesthetized animals for the passage of the drug of interest to be evaluated. By maintaining innervation and blood flow preserved, these absorption models keep a faithful reproduction of what happens in vivo [110]. On the other hand, ex situ models are those performed outside the original place. For comparative purposes, in a study with bacterial cellulose used for tissue engineering cultures, the bacteria were cultured with in situ and ex situ modifications. Culture of bacterial cellulose in situ were characterized by adding in the medium other material such as an additive, while in the ex situ culture modification bacteria just received chemical treatment or absorption of other materials after the bacteria membranes have been formed in culture [114]. However, probably the most relevant ex situ method for biomedical sciences is the ex situ perfusion for organ preservation, which can restore circulation to regain the organ function (e.g. liver) prior to transplantation [115-117].
Ex situ techniques are also described in quality control of immunobiological products and in biodiversity conservation. In vaccine analysis, we find particle size distribution analysis, zeta potential and settling rate that are performed outside pre-filled syringes [118]. In biodiversity, the ex situ is used to describe gene banks, captive breeding, and botanical garden for biological diversity conservation. These methods are an essential part for the reintroduction of endangered species and have some restrictions such as high cost, employee demand and energy resources [119,120]. Examples of in situ and ex situ models could be seen in Figure 3.
In Vivo
During human history and development, humans always took advantage from numerous scientific researches involving animals. Virtually, from the past century, every drug released on the market was dependent on the use of in vivo models in some step of their development [121].
The progress of specific and efficacious treatments of diabetes, several types of cancer, heart surgery, among others, were only possible using animals in scientific research [122]. A recent survey demonstrated that 44% of the population and more than 80% of medical students support the use of animals in research, although they also state that alternative methods should be applied, whenever possible, in order to reduce the number of animals for that purpose [123].
Overall, researchers seek to investigate organisms at multiple levels, starting from their molecules, to cells, tissues, organs, and up to their systems, both in disease and health conditions. For most of them, there is the option to conduct studies using in silico or in vitro models, such as molecular modeling and cell line culture, respectively. Cell line culture, specifically, has largely evolved in the past 20 years, and became a usual approach to mimic the complex structures of tissues, currently playing key roles in research. In this regard, several animal studies can be replaced due to the advent of such technology [124,125]. Nonetheless, the understanding of physiological processes and systemic interactions still requires in vivo experiments.
In vivo model (i.e. within the living) describes a methodology where the whole subject is studied while alive. Although in vivo models can be performed with non-animals, such as plants and seeds [126], in vivo experiments using animal models are the most used, reaching more than 100 million animal experiments per year in the world (e.g. Great Britain performed 1,078,738 experimental procedures just in mice in 2018) [127,128]. In vivo animal assays are carried out when specific situations are not suitable to be done in humans, due to a possible risk to their physical and/or physiological integrity. The animal use in research is not only due to their similarity with human physiology, but also because human diseases may also affect other animal species [129]. Additionally, it is possible to have in vivo models with specific mutations that directly cause or strongly predispose the animal to a desired anomaly or disease, being referred to as “experimental models of a disease” [129-132].
Research with animals have the purpose of providing experimental data in one living organism to study a phenomenon in another species, aiming to obtain preclinical data to determine how a hypothesis would work on humans [133]. On the other hand, the results obtained using animals are not necessarily reproducible in humans. Despite great similarities that a species may have with humans, genetic differences such as gene families, redundancies, and regulation of gene expression patterns may influence the variable responses [122].
Although massive worldwide investments in drug development, the overall success rate of new drugs, especially on clinical trials, remains low. One possible explanation relies on the preclinical research phase, when ‘inappropriate animal models’ are chosen because of lacking a predictive model, which leads to nonextrapolating conclusions [134]. In this context, it is fundamental to select the correct animal model according to the objectives of the proposed research [134].
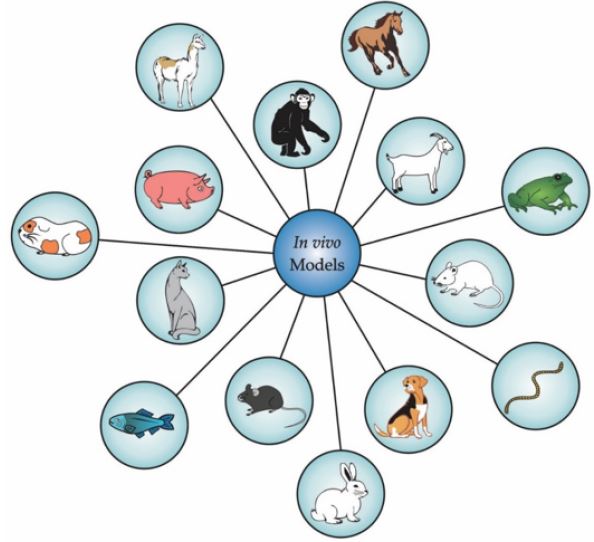
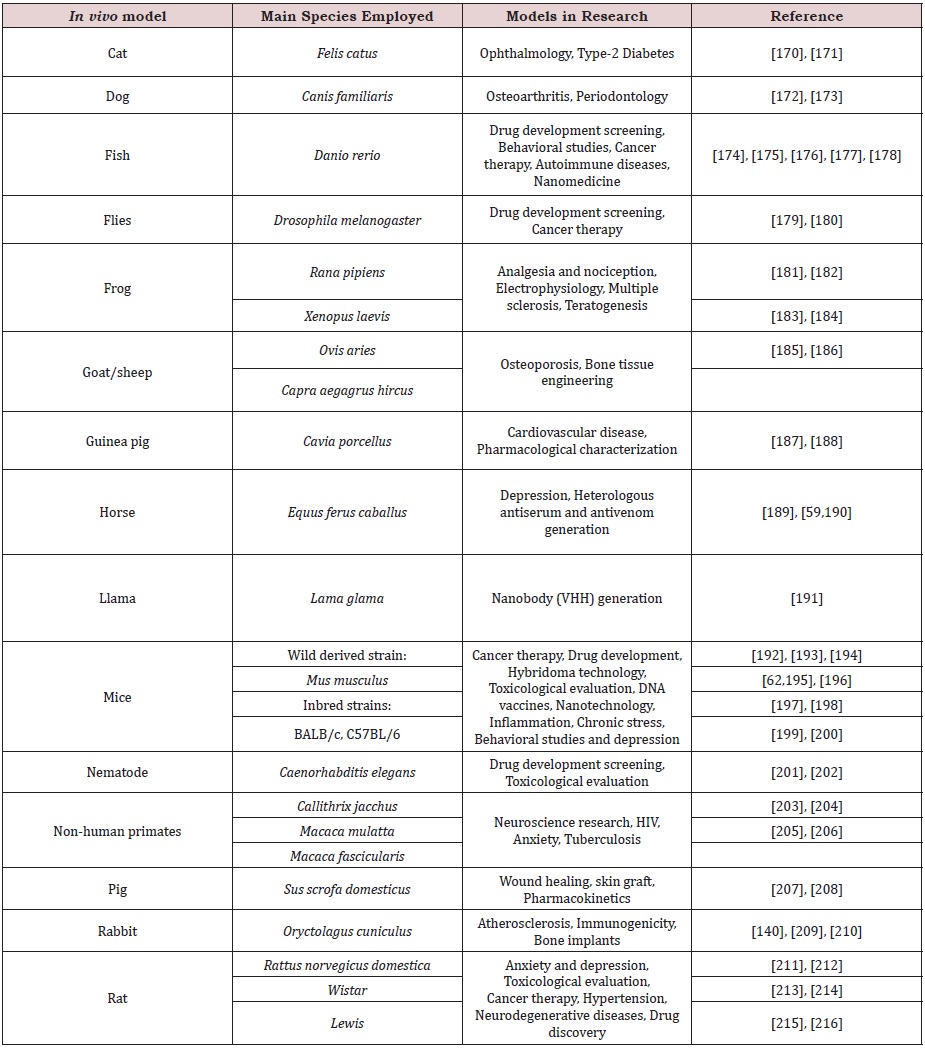
Although rodents (rats, mice, and guinea pigs) are mostly used due to their shorter lifespans, which creates the possibility of producing many generations [135], there are many other available animal models, such as insects (Drosophila ssp.) [136], fish (Danio rerio, or zebrafish) [137], nematodes (Caenorhabditis elegans) [138], frogs (Xenopus ssp.) [139], rabbits [140], horses [141], dogs [142], cats [143], pigs [144], and even non-human primate models [145], among others (Figure 4). Some examples of preclinical animal models and their use in research are listed in Table 1.
Non-human primates have been extensively used as models in animal research, due to their close phylogenetic relationship to humans, with validated similarities in terms of behavioral and biochemical activities, as well as gene expression patterns [146]. Based on that, some non-human primates are considered key in vivo models for specific research fields, including studies with the acquired immunodeficiency disease syndrome (AIDS), autoimmune diseases, Parkinson’s disease, hepatitis, diabetes, physiological and psychiatric disorders, transplants, toxicological effects, dentistry, drug abuse and vaccine development [133,147].
Nevertheless, there are many regulatory requirements and policies to perform experiments on non-human primates, especially because they are large animals, intelligent, social, long-lived, and non-domesticated animals [148]. Therefore, non-human primates should only be used in specific cases, when the less-sentient species do not meet the requirements of the research.
PICKING THE BEST RESEARCH MODEL
Selecting the best research model can be a real challenge. Based on the efforts to 3Rs principle (Reduce, Refine and Replace the use of animals), many in vitro and in silico models have been explored as alternatives to in vivo methods, such as organs-ina- chip and toxicity prediction, respectively. Nevertheless, these alternative models should be mechanistically based on the in vivo process, scientifically supported, and based on well-known responses described in vivo, which means that the model should be validated before being adopted. Fortunately, the availability of in vivo alternative tests has increased intensely, and many are very attractive in cost and time [149].
Researchers can obtain information about the alternative validated methods through different regulatory agencies [150-152]. Based on the above, method validation is vital to check the safety and efficacy of the research. In concern to the biopharmaceutical drugs, many regulatory organizations have addressed this issue in the chemical and pharmaceutical industry. For instance, the analytic validation can be assessed at two levels.
1. The pre-study validation that aims to show the method can achieve its objectives; and
2. In-study validation that verifies if the method remains valid over time by including quality control samples in routine runs [153,154].
However, the main challenge in biomedical sciences is when the model cannot be compared to humans. Animal models have supported critical advances in biomedical research, offering deep insights into several diseases. But they have been less successful as a basis for advancing human health as the failure of translation from animal models to human patients has often been disappointing [155].
A common criticism of model-based biomedical research is that while we have gotten very good at curing mouse models, we have made much less progress for human patients and, although the assumption that the animal models are a good proxy for humans is a central tenet of biomedical research, it is not always a reliable one [156-158]. For instance, it is well-known the gap between human and murine inflammation responses. Thus, how can we compare mice models of human immune-related diseases (e.g. diabetes, asthma, and multiple sclerosis)? There are many animal data failing to predict human responses to potential immunological therapies [159].
Choosing of the best experiment to validate a hypothesis is not the only issue for researchers. There is no consensus in the scientific community for the model classification. Misunderstandings with in vitro, ex vivo, in situ, ex situ, and in vivo models are very frequent, simply because there is no article or review giving satisfactory information about differentiating these models. Although most academy agrees that in vivo models are those conducted in animals, the literature presents articles in which assays conducted out of animals (e.g. ex vivo and in vitro) are referred as in vivo. For instance, the chick chorioallantoic membrane (CAM) assay can be defined by researchers either as an ex vivo [160] or as an in vivo [161] approach.
However, since the assay is developed in the alive chick embryo (which last 21 days to develop), should fit better to classify the assay as an in vivo model [162].
Furthermore, it is easy to come across hundreds of research articles, even in highly reputed journals, where cell line culture experiments are referred to as both in vitro, and sometimes, in vivo [163-166]. Thus, which research model should be the best to classify cultures performed with cell lines? Following the previous definition, we encourage researchers to refer as in vitro all experiments developed with cell lines. Histological studies also easily generate confusion among researchers since they can be classified as in vivo [167], ex vivo [168], as well as in vitro [169] models. Basically, the differences between the models are based on the way that the tissue was obtained. Ex vivo and in vitro histology should follow Sections. On the other hand, in vivo histology is a peculiar model, where the tissue is accessed through an intravascular ultrasound radiofrequency technique, named also virtual histology.
CONCLUSION
As can be seen above, researchers can get very confused to pick and classify their research models since there are no guidelines for choosing and classifying them so far. In addition, these models and their classification may vary considerably among the several scientific fields, different research organizations, and individual investigators. This review is the first to clarify the main definitions of the principal research models used in biomedical sciences with marked examples, aiming to guide and align researchers during their experimental practices.
ACKNOWLEDGEMENT
We thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo Research Foundation; scholarships to ISO no. 2017/03580-9, ELPJ no. 2016/04761-4, and FAC no. 2017/14035-1), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, The National Council for Scientific and Technological Development, scholarship to MBP no. 307155/2017- 0), and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES, Finance Code 001, scholarship to IGF). Conselho Nacional de Desenvolvimento Científico e Tecnológico: scholarships to MBP no. 307155/2017 and FAC no. 155276/2018- 2. Coordenação de Aperfeiçoamento de Pessoal de Nível Superior- Brasil: scholarships to ELPJ no. 88881.186830/2018-01 and IGF.
AUTHORS CONTRIBUTION
ISO, GMAS, FAC, ELPJ, IGF and UZ wrote the review, FAC provided figures and wrote the review and MBP was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
REFERENCES
- Marecková E, Simon F, Cervený L (2002) Latin as the language of medical terminology: some remarks on its role and prospects. Swiss Med Wkly 132: 581-587.
- Brunton L (1916) Latin as a universal language. Nature 96: 649.
- Bieliaieva OM, Lysanets YV, Znamenska IV, Rozhenko IV, Nikolaieva NM (2017) Terminological collocations in medical latin and english: a comparative study. Wiad Lek 70: 139-143.
- Soeiro M de NC, de Souza EM, da Silva CF, Batista D da GJ, Batista MM, et al. (2013) In Vitro and In Vivo studies of the antiparasitic activity of sterol 14α-demethylase (CYP51) Inhibitor VNI against drug-resistant strains of trypanosoma cruzi. Antimicrob Agents Chemother 57: 4151-4163.
- Young VB, Knox KA, Pratt JS, Cortez JS, Mansfield LS, et al. (2004) In Vitro and In Vivo Characterization of helicobacter hepaticus cytolethal distending toxin mutants. Infection and Immunity. 72(5): 2521-2527.
- Becker OM, Marantz Y, Shacham S, Inbal B, Heifetz A, et al. (2004) G protein-coupled receptors: In silico drug discovery in 3D. PNAS 101(31): 11304-11309.
- Kampmann T, Yennamalli R, Campbell P, Stoermer MJ, Fairlie DP, et al. (2009) In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral Research 84(3): 234-241.
- Ahmed V, Opoku A, Aziz Z (2016) Research methodology in the built environment: A selection of case studies. Routledge, New York, USA.
- Amano T, González-Varo JP, Sutherland WJ (2016) Languages are still a major barrier to global science. PLoS biology 14: e2000933-e2000933.
- Scotti L, Ghasemi J, Scotti MT (2018) Editorial: In Silico methodologies applied to drug discovery. CCHTS 21(3): 150-151.
- Miramontes Pedro (1989) DNA and RNA physicochemical constraints, cellular automata and molecular evolution. Los Alamos, New Mexico, USA.
- Colquitt RB, Colquhoun DA, Thiele RH (2011) In silico modelling of physiologic systems. Best practice & research Clinical anaesthesiology 25(4): 499-510.
- Amberg A (2013) In Silico Methods. In: Vogel HG, et al. (Eds). Drug discovery and evaluation: safety and pharmacokinetic assays. Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 1273-1296.
- Ekins S, Mestres J, Testa B (2007) In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol 152(1): 9-20.
- Cavaliere F, Cozzini P (2018) New in Silico trends in food toxicology. Chem Res Toxicol 31(10): 992-993.
- Scior T, Bender A, Tresadern G, Medina-Franco JL, Martínez-Mayorga K, et al. (2012) Recognizing pitfalls in virtual screening: A Critical Review. J Chem Inf Model 52(4): 867-881.
- Marchal T (2015) In Vivo, In Vitro, In Silico! Best Practices.
- Trisilowati, Mallet DG (2012) In Silico experimental modeling of cancer treatment. ISRN Oncology 2012: 828701.
- Yan G, Wang X, Chen Z, Wu X, Pan J, et al. (2017) In-silico ADME studies for new drug discovery: from chemical compounds to Chinese herbal medicines. Curr Drug Metab 18(6): 535-539.
- Sotomayor M, Schulten K (2007) Single-molecule experiments in Vitro and in Silico. Science 316(5828): 1144-1148.
- Fischer HP (2008) Mathematical modeling of complex biological systems. Alcohol Res Health 31(1): 49-59.
- Sethi A, Joshi K, Sasikala K, Alvala M (2019) Molecular docking in modern drug discovery: principles and recent applications. drug discovery and development - New Advances.
- Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE (1982) A geometric approach to macromolecule-ligand interactions. J Mol Biol 161(2): 269- 288.
- Pagadala NS, Syed K, Tuszynski J (2017) Software for molecular docking: a review. Biophys Rev 9(2): 91-102.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3): 403-410.
- Marti-Renom MA, Madhusudhan MS, Sali A (2004) Alignment of protein sequences by their profiles. Protein Sci 13(4): 1071-1087.
- Pucca MB, Cerni FA, Peigneur S, Bordon KCF, Tytgat J, et al. (2015) Revealing the function and the structural model of Ts4: Insights into the “Non-Toxic” Toxin from Tityus serrulatus Venom. Toxins (Basel) 7(7): 2534-2550.
- Bridge LJ, Mead J, Frattini E, Winfield I, Ladds G (2018) Modelling and simulation of biased agonism dynamics at a G protein-coupled receptor. J Theor Biol 442: 44-65.
- Myatt GJ, Ahlberg E, Akahori Y, Allen D, Amberg A, et al. (2018) In silico toxicology protocols. Regulatory Toxicology and Pharmacology 96: 1-17.
- Idakwo G, Luttrell J, Chen M, Hong H, Zhou Z, et al. (2018) A review on machine learning methods for in silico toxicity prediction. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 36(4): 169-191.
- Kamble A, Srinivasan S, Singh H (2019) In-Silico bioprospecting: Finding better enzymes. Mol Biotechnol 61(1): 53-59.
- Roy A, Nair S, Sen N, Soni N, Madhusudhan MS (2017) In silico methods for design of biological therapeutics. Methods 131: 33-65.
- Pappalardo F, Russo G, Tshinanu FM, Viceconti M (2019) In silico clinical trials: concepts and early adoptions. Brief Bioinform 20(5): 1699-1708.
- Brown D, Namas RA, Almahmoud K, Zaaqoq A, Sarkar J, et al. (2015) Trauma in silico: Individual-specific mathematical models and virtual clinical populations. Science Translational Medicine 7: 285ra61- 285ra61.
- Pearson RM (1986) In-vitro techniques: can they replace animal testing? Hum Reprod 1(8): 559-560.
- Weil H, Willams TI (1950) History of chromatography. Nature 166: 1000.
- Iler RK (1979) Wiley-interscience publication. The chemistry of silica solubility, polymerisation, colloid and surface properties, and biochemistry. New York, USA.
- Lehninger AL, Nelson DL, Cox MM (2013) Lehninger principles of biochemistry, (6th edn), WH Freeman, New York, USA.
- Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C Content of deoxyribonucleic acid by high-performance liquid chromatography. International Journal of Systematic Bacteriology 39(2): 159-167.
- Maher S, Jjunju FPM, Taylor S (2015) Colloquium: 100 years of mass spectrometry: Perspectives and future trends. Reviews of Modern Physics 87: 113-135.
- Lazzari E, Arena K, Caramão EB, Herrero M (2019) Quantitative analysis of aqueous phases of bio-oils resulting from pyrolysis of different biomasses by two-dimensional comprehensive liquid chromatography. Journal of Chromatography A 1602: 359-367.
- Morsa D, Baiwir D, La Rocca R, Zimmerman TA, Hanozin E, et al. (2019) Multi-enzymatic limited digestion: The next-generation sequencing for proteomics? J Proteome Res 18(6): 2501-2513.
- O’Connell TX, Horita TJ, Kasravi B (2005) Understanding and interpreting serum protein electrophoresis. Am Fam Physician 71: 105-112.
- Tiselius A (1937) A new apparatus for electrophoretic analysis of colloidal mixtures. Trans Faraday Soc 33: 524-531.
- Hames BD (1998) Gel electrophoresis of proteins: A practical approach. OUP Oxford.
- Kunkel HG, Tiselius A (1951) Electrophoresis of proteins on filter paper. J Gen Physiol 35(1): 89-118.
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem 203(1): 173-179.
- Yao H, Wynendaele E, Xu X, Kosgei A, De Spiegeleer B (2018) Circular dichroism in functional quality evaluation of medicines. J Pharm Biomed Anal 147: 50-64.
- Bonjoch NP, Tamayo PR (2003) Protein content quantification by bradford method. In: Reigosa Roger MJ, (Ed.) Handbook of Plant Ecophysiology Techniques. Dordrecht: Kluwer Academic Publishers, pp. 283-295.
- Peterson GL (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Analytical Biochemistry 83(2): 346-356.
- McREE DE (1999) Practical protein crystallography. Elsevier.
- Valério AA, Corradini AC, Panunto PC, Mello SM, Hyslop S (2002) Purification and characterization of a phosphodiesterase from bothrops alternatus snake venom. J Protein Chem 21(8): 495-503.
- Bjork W (1963) Purification of phosphodiesterase from Bothrops atrox venom, with special consideration of the elimination of monophosphatases. J Biol Chem 238: 2487-2490.
- Di Ferrante N (1956) Turbidimetric measurement of acid mucopolysaccharides and hyaluronidase activity. J Biol Chem 220(1): 303-306.
- Pukrittayakamee S, Warrell DA, Desakorn V, McMichael AJ, White NJ, et al. (1988) The hyaluronidase activities of some Southeast Asian snake venoms. Toxicon 26(7): 629-637.
- Selvanayagam ZE, Gopalakrishnakone P (1999) Tests for detection of snake venoms, toxins and venom antibodies: review on recent trends (1987-1997). Toxicon 37(4): 565-586.
- Piliarik M, Vaisocherová H, Homola J (2009) Surface plasmon resonance biosensing. Methods Mol Biol 503: 65-88.
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, et al. (1986) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51 (Pt 1): 263-273.
- Pucca MB, Cerni FA, Janke R, Bermúdez ME, Ledsgaard L, et al. (2019) History of envenoming therapy and current perspectives. Front Immunol.
- Ryazantsev DY, Voronina DV, Zavriev SK (2016) Immuno-PCR: achievements and perspectives. Biochemistry Mosc 81(13): 1754-1770.
- Dal Ferro M, Rizzo S, Rizzo E, Marano F, Luisi I, et al. (2019) Phage display technology for human monoclonal antibodies. Methods Mol Biol 1904: 319-338.
- Laustsen AH, Karatt-Vellatt A, Masters EW, Arias AS, Pus U, et al. (2018) In vivo neutralization of dendrotoxin-mediated neurotoxicity of black mamba venom by oligoclonal human IgG antibodies. Nature Communications 9(1): 3928.
- Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, Laustsen AH (2018) Basics of antibody phage display technology. Toxins (Basel) 10(6): 236.
- Karahalil B (2016) Overview of systems biology and omics technologies. Curr Med Chem 23(37): 4221-4230.
- Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, et al. (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4(7): 844-847.
- Zhang D, Salto TM, Putti TC, Do E, Koay ESC (2003) Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 16: 79-84.
- Jedrzejczak-Silicka M (2017) History of cell culture. New insights into cell culture technology.
- Horrocks C, Halse R, Suzuki R, Shepherd PR (2003) Human cell systems for drug discovery. Curr Opin Drug Discov Devel 6(4): 570-575.
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, et al. (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603-607.
- Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, et al. (2019) Next-generation characterization of the cancer cell line encyclopedia. Nature 569: 503-568.
- Risbridger GP (2015) Human cell lines as tools of our trade: “Laying It on the (Cell) Line.” Mol Endocrinol 29(1): 1-2.
- Prinsen MK, Koëter HB (1993) Justification of the enucleated eye test with eyes of slaughterhouse animals as an alternative to the draize eye irritation test with rabbits. Food Chem Toxicol 31(1): 69-76.
- Prinsen MK (1996) The chicken enucleated eye test (CEET): a practical (pre)screen for the assessment of eye irritation/corrosion potential of test materials. Food Chem Toxicol 34(3): 291-296.
- Mummery C, van de Stolpe A, Roelen BAJ, Clevers H (2013) Chapter 13 - Human Stem Cells for Organs-on-Chips: Clinical Trials Without Patients?11Part of this chapter was adapted from an Organs-on-Chips meeting report, Lab on a Chip 2013, DOI: 10.1039/c3lc50248a. Stem Cells (2nd Edn). Boston: Academic Press, p. 343-361.
- Hodsden S (2015) Human organ-mimicking chip could revolutionize clinical trials. Med Device Online.
- Martins M, Schelz Z, Martins A, Molnar J, Hajös G, et al. (2007) In vitro and ex vivo activity of thioridazine derivatives against Mycobacterium tuberculosis. International Journal of Antimicrobial Agents 29(3): 338- 340.
- Saeidnia S, Manayi A, Abdollahi M (2015) From in vitro experiments to in vivo and clinical studies; pros and cons. Curr Drug Discov Technol 12(4): 218-224.
- Clift MJD, Gehr P, Rothen-Rutishauser B (2011) Nanotoxicology: a perspective and discussion of whether or not in vitro testing is a valid alternative. Arch Toxicol 85(7): 723-731.
- Griesinger C, Desprez B, Coecke S, Casey W, Zuang V (2016) Validation of alternative In Vitro methods to animal testing: Concepts, challenges, processes and tools. Adv Exp Med Biol 856: 65-132.
- Tacken PJ, de Vries IJM, Torensma R, Figdor CG (2007) Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat Rev Immunol 7(10): 790-802.
- Dusinska M, Rundén-Pran E, Schnekenburger J, Kanno J (2017) Toxicity tests: In Vitro and In Vivo. adverse effects of engineered nanomaterials. Elsevier 51-82.
- Parente RJA, Tomazett MV, Pigosso LL, Bailão AM, Ferreira de Souza A, et al. (2018) In vitro, ex vivo and in vivo models: A comparative analysis of Paracoccidioides spp. proteomic studies. Fungal Biology 122(6): 505- 513.
- Gregory EK, MA Emran Bashar, Tan M (2012) Ex Vivo gene therapy and vision. CGT 12(2): 103-115.
- Snijder B, Vladimer GI, Krall N, Miura K, Schmolke AS, et al. (2017) Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: Interim results from a single-arm, openlabel, pilot study. The Lancet Haematology 4(12): e595-e606.
- Gowing G, Svendsen S, Svendsen CN (2017) Ex vivo gene therapy for the treatment of neurological disorders. Prog Brain Res 230: 99-132.
- Granzin M, Wagner J, Köhl U, Cerwenka A, Huppert V, et al. (2017) Shaping of natural killer cell antitumor activity by Ex Vivo cultivation. Front Immunol 8: 458.
- Kumar S, Geiger H (2017) HSC niche biology and hsc expansion Ex vivo. Trends Mol Med 23(9): 799-819.
- Zotti A, Banzato T, Gelain ME, Centelleghe C, Vaccaro C, et al. (2015) Correlation of renal histopathology with renal echogenicity in dogs and cats: an ex-vivo quantitative study. BMC Veterinary Research 11: 99.
- Chung WY, Wanford JJ, Kumar R, Isherwood JD, Haigh RD, et al. (2019) An ex vivo porcine spleen perfusion as a model of bacterial sepsis. ALTEX 36(1): 29-38.
- Schraufnagel DP, Steffen RJ, Vargo PR, Attia T, Elgharably H, et al. (2018) Devices for ex vivo heart and lung perfusion. Expert Rev Med Devices 15(3): 183-191.
- Naldini L (2011) Ex vivo gene transfer and correction for cell-based therapies. Nat Rev Genet 12(5): 301-315.
- OECD. Test No. 437: Bovine corneal opacity and permeability test method for identifying i) chemicals inducing serious eye damage and ii) Chemicals not requiring classification for eye irritation or serious eye damage. OECD 2017.
- Wilson SL, Ahearne M, Hopkinson A (2015) An overview of current techniques for ocular toxicity testing. Toxicology 327: 32-46.
- Van den Berghe C, Guillet MC, Compan D (2005) Performance of porcine corneal opacity and permeability assay to predict eye irritation for water-soluble cosmetic ingredients. Toxicology in Vitro 19(6): 823-830.
- Gómez-Sintes R, Villarejo-Zori B, Serrano-Puebla A, Esteban-Martínez L, Sierra-Filardi E, et al. (2017) Standard assays for the study of autophagy in the Ex Vivo Retina. Cells 6(4): 37.
- Kooragayala K, Gotoh N, Cogliati T, Nellissery J, Kaden TR, et al. (2015) Quantification of oxygen consumption in retina Ex Vivo demonstrates limited reserve capacity of photoreceptor mitochondria. Invest Ophthalmol Vis Sci 56(13): 8428-8436.
- Gotlieb N, Rosenne E, Matzner P, Shaashua L, Sorski L, et al. (2015) The misleading nature of in vitro and ex-vivo findings in studying the impact of stress hormones on NK cell cytotoxicity. Brain Behav Immun 45: 277- 286.
- Oishi N, Montes-Moreno S, Feldman AL (2018) In situ neoplasia in lymph node pathology. Semin Diagn Pathol 35(1): 76-83.
- Edlefsen KL, Greisman HA, Yi HS, Mantei KM, Fromm JR (2011) Early lymph node involvement by mantle cell lymphoma limited to the germinal center: report of a case with a novel “follicular in situ” growth pattern. Am J Clin Pathol 136(2): 276-281.
- Langer-Safer PR, Levine M, Ward DC (1982) Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci U S A 79(14): 4381-4385.
- Huber D, Voith von Voithenberg L, Kaigala GV (2018) Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro and Nano Engineering 1: 15-24.
- Onozato ML, Yapp C, Richardson D, Sundaresan T, Chahal V, et al. (2019) Highly Multiplexed Fluorescence in Situ Hybridization for in Situ Genomics. J Mol Diagn 21(3): 390-407.
- Nguyen HT, Dupont LN, Cuttaz EA, Jean AM, Trouillon R, et al. (2018) Breast cancer HER2 analysis by extra-short incubation microfluidics-assisted fluorescence in situ hybridization (ESIMA FISH). Microelectronic Engineering 189: 33-38.
- El-Menoufy MAM, Mourad ZI, Farahat NM (2018) The prognostic impact of loss of chromosome 7 material detected by fluorescence in situ hybridization (FISH) in myeloid malignancies. J Egypt Natl Canc Inst 30(4): 133-138.
- Chang AE, Ganz PA, Hayes DF, Kinsella TJ, Pass HI, et al. (2006) Oncology: an evidence-based approach. Springer Science Business Media Inc: Springer e-books, New York, USA.
- Parikh U, Chhor CM, Mercado CL (2018) Ductal carcinoma in situ: The whole truth. AJR Am J Roentgenol 210(2): 246-255.
- Karube K, Scarfò L, Campo E, Ghia P (2014) Monoclonal B cell lymphocytosis and “in situ” lymphoma. Semin Cancer Biol 24: 3-14.
- Thomas JA, Buchsbaum RN, Zimniak A, Racker E (1979) Intracellular pH measurements in Ehrlich ascites tumor cells utilizing spectroscopic probes generated in situ. Biochemistry 18(11): 2210-2218.
- Kunishige JH, Doan L, Brodland DG, Zitelli JA (2019) Comparison of surgical margins for lentigo maligna versus melanoma in situ. J Am Acad Dermatol 81(1): 204-212.
- Stappaerts J, Brouwers J, Annaert P, Augustijns P (2015) In situ perfusion in rodents to explore intestinal drug absorption: Challenges and opportunities. Int J Pharm 478(2): 665-681.
- Lozoya-Agullo I, Zur M, Fine-Shamir N, Markovic M, Cohen Y, et al. (2017) Investigating drug absorption from the colon: Single-pass vs. Doluisio approaches to in-situ rat large-intestinal perfusion. Int J Pharm 527(1-2): 135-141.
- Luo Z, Liu Y, Zhao B, Tang M, Dong H, et al. (2013) Ex vivo and in situ approaches used to study intestinal absorption. J Pharmacol Toxicol Methods 68(2): 208-216.
- Yang H, Zhai B, Fan Y, Wang J, Sun J, et al. (2018) Intestinal absorption mechanisms of araloside A in situ single-pass intestinal perfusion and in vitro Caco-2 cell model. Biomedicine & Pharmacotherapy 106: 1563-1569.
- Stumpf TR, Yang X, Zhang J, Cao X (2018) In situ and ex situ modifications of bacterial cellulose for applications in tissue engineering. Materials Science and Engineering: C 82: 372-383.
- Bral M, Gala-Lopez B, Bigam DL, Freed DH, Shapiro AMJ (2018) Ex situ liver perfusion: Organ preservation into the future. Transplantation Reviews 32(3): 132-141.
- Liu Q, Nassar A, Buccini L, Grady P, Soliman B, et al. (2018) Ex situ 86- hour liver perfusion: Pushing the boundary of organ preservation. Liver Transplantation 24(4): 557-561.
- Werner NL, Alghanem F, Rakestraw SL, Sarver DC, Nicely B, et al. (2017) Ex Situ Perfusion of Human Limb Allografts for 24 Hours. Transplantation 101(3): e68-e74.
- Lewis LM, Guo J, Torres E, Wang J, Billones H, Kolhe P, et al. (2017) Ex Situ and In Situ Characterization of vaccine suspensions in pre-filled syringes. Journal of Pharmaceutical Sciences 106(8): 2163-2167.
- Mounce R, Smith P, Brockington S (2017) Ex situ conservation of plant diversity in the world’s botanic gardens. Nature Plants 3: 795-802.
- Kasso M, Balakrishnan M (2013) Ex Situ conservation of biodiversity with particular emphasis to Ethiopia. International Scholarly Research Notices p.11.
- Worker T (2004) The use of non-human animals in research: A guide for scientists. Alternatives to Laboratory Animals 32: 119-120.
- Barré SF, Montagutelli X (2015) Animal models are essential to biological research: Issues and perspectives. Future Sci OA 1(4): FSO63.
- Joffe AR, Bara M, Anton N, Nobis N (2016) The ethics of animal research: a survey of the public and scientists in North America. BMC Medical Ethics 17: 17.
- Chen FM, Liu X (2016) Advancing biomaterials of human origin for tissue engineering. Progress in Polymer Science 53: 86-168.
- Zhang YS, Yue K, Aleman J, Moghaddam KM, Bakht SM, Yang J, et al. (2017) 3D Bioprinting for Tissue and Organ Fabrication. Ann Biomed Eng 45(1): 148-163.
- Kumari M, Ernest V, Mukherjee A, Chandrasekaran N (2012) In vivo nanotoxicity assays in plant models. Methods Mol Biol 926: 399-410.
- Ebel I (2013) Pesquisa usa 115 milhões de animais por ano no mundo, diz ativista.
- UAR news team (2019). Animal research numbers in 2018. Understanding animal research.
- Perlman RL (2016) Mouse models of human disease: An evolutionary perspective. Evol Med Public Health 2016(1): 170-176.
- Council NR (2004) Science, medicine and animals.
- Guan C, Ye C, Yang X, Gao J (2010) A review of current large-scale mouse knockout efforts. Genesis 48(2): 73-85.
- Kim IY, Shin JH, Seong JK (2010) Mouse phenogenomics, toolbox for functional annotation of human genome. BMB Rep 43: 79-90.
- Andersen ML, Winter LMF (2019) Animal models in biological and biomedical research - experimental and ethical concerns. An Acad Bras Cienc 91(suppl 1): e20170238.
- Denayer T, Stöhr T, Van Roy M (2014) Animal models in translational medicine: Validation and prediction. New Horizons in Translational Medicine 2(1): 5-11.
- Vandamme TF (2015) Rodent models for human diseases. Eur J Pharmacol 759: 84-89.
- Alaraby M, Annangi B, Marcos R, Hernández A (2016) Drosophila melanogaster as a suitable in vivo model to determine potential side effects of nanomaterials: A review. J Toxicol Environ Health B Crit Rev 19(2): 65-104.
- Dooley K, Zon LI (2000) Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev 10(3): 252-256.
- Kaletta T, Hengartner MO (2006) Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 5(5): 387-398.
- Nutt LK (2012) The Xenopus oocyte: a model for studying the metabolic regulation of cancer cell death. Semin Cell Dev Biol 23 :412-418.
- Fan J, Chen Y, Yan H, Niimi M, Wang Y, Liang J (2018) Principles and applications of rabbit models for atherosclerosis research. J Atheroscler Thromb 25(3): 213-220.
- Raweerith R, Ratanabanangkoon K (2005) Immunochemical and biochemical comparisons of equine monovalent and polyvalent snake antivenoms. Toxicon 45(3): 369-375.
- Berglundh T, Abrahamsson I, Lang NP, Lindhe J (2003) De novo alveolar bone formation adjacent to endosseous implants. Clin Oral Implants Res 14(3): 251-262.
- Topolnik L, Steriade M, Timofeev I (2003) Hyperexcitability of intact neurons underlies acute development of trauma-related electrographic seizures in cats in vivo. Eur J Neurosci 18(3): 486-496.
- Barbero AM, Frasch HF (2009) Pig and guinea pig skin as surrogates for human in vitro penetration studies: a quantitative review. Toxicol In Vitro 23(1): 1-13.
- Machado CJ, Bachevalier J (2003) Non-human primate models of childhood psychopathology: the promise and the limitations. J Child Psychol & Psychiat 44(1): 64-87.
- Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, et al. (2014) Why primate models matter. Am J Primatol 76(9): 801-827.
- Bontrop RE (2001) Non-human primates: essential partners in biomedical research. Immunological Reviews 183: 5-9.
- Tardif SD, Coleman K, Hobbs TR, Lutz C (2013) IACUC review of nonhuman primate research. ILAR J 54(2): 234-245.
- (2017) National academies of sciences, Studies D on E and L, Toxicology B on ES and Evaluations C on 21st CS into R-B. Model and assay validation and acceptance. National Academies Press USA.
- O’Connor L (2017) EU reference laboratory for alternatives to animal testing. EU Science Hub-European Commission, Belgium.
- (2019) AltTox.org. animal testing alternatives | Non-Animal Methods.
- NIEHS (2019) Interagency coordinating committee on the validation of alternative methods.
- Stöckl D, D’Hondt H, Thienpont LM (2009) Method validation across the disciplines--critical investigation of major validation criteria and associated experimental protocols. J Chromatogr B Analyt Technol Biomed Life Sci 877(23): 2180-2190.
- Oliva A, Fariña JB, Llabrés M (2016) Pre-study and in-study validation of a size-exclusion chromatography method with different detection modes for the analysis of monoclonal antibody aggregates. J Chromatogr B Analyt Technol Biomed Life Sci 1022: 206-212.
- van der Staay FJ, Arndt SS, Nordquist RE (2009) Evaluation of animal models of neurobehavioral disorders. Behav Brain Funct 5: 11.
- Pound P, Ebrahim S, Sandercock P, Bracken MB, Roberts I (2004) Where is the evidence that animal research benefits humans? BMJ 328(7438): 514-517.
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA 110(9): 3507-3512.
- Ioannidis JPA (2012) Extrapolating from animals to humans. Sci Transl Med 4(151): 151ps15.
- Mestas J, Hughes CCW (2004) Of mice and not men: differences between mouse and human immunology. J Immunol 172(5): 2731- 2738.
- Jedelská J, Strehlow B, Bakowsky U, Aigner A, Höbel S, et al. (2013) The chorioallantoic membrane assay is a promising ex vivo model system for the study of vascular anomalies. In Vivo 27(6): 701-705.
- Lokman NA, Elder ASF, Ricciardelli C, Oehler MK (2012) Chick Chorioallantoic Membrane (CAM) assay as an In Vivo model to study the effect of newly identified molecules on ovarian cancer invasion and metastasis. Int J Mol Sci 13: 9959-9970.
- Ribatti D (2010) The chick embryo chorioallantoic membrane in the study of angiogenesis and metastasis: The CAM assay in the study of angiogenesis and metastasis. Springer Science & Business Media, Germany.
- Antoni D, Burckel H, Josset E, Noel G (2015) Three-dimensional cell culture: A breakthrough in Vivo. Int J Mol Sci 16(3): 5517-5527.
- Lelièvre SA, Kwok T, Chittiboyina S (2017) Architecture in 3D cell culture: An essential feature for in vitro toxicology. Toxicol In Vitro 45(3): 287-295.
- Kutscher HL, Morse GD, Prasad PN, Reynolds JL (2019) In vitro pharmacokinetic cell culture system that simulates physiologic nanoparticle exposure to macrophages. Pharm Res 36(3): 44.
- Allen DD, Caviedes R, Cárdenas AM, Shimahara T, Segura AJ, et al. (2005) Cell lines as in vitro models for drug screening and toxicity studies. Drug Dev Ind Pharm 31(8): 757-768.
- Nasu K, Tsuchikane E, Katoh O, Vince DG, Virmani R, et al. (2006) Accuracy of in vivo coronary plaque morphology assessment. Journal of the American College of Cardiology 47(12): 2405-2412.
- Li L, Pahwa S, Penzias G, Rusu M, Gollamudi J, et al. (2017) Co- Registration of ex vivo Surgical Histopathology and in vivo T2 weighted MRI of the Prostate via multi-scale spectral embedding representation. Sci Rep 7(1): 8717.
- Alam K, Al Ghaithi A, Piya S, Saleem A (2019) In-vitro experimental study of histopathology of bone in vibrational drilling. Med Eng Phys 67: 78-87.
- Narfström K, Holland DK, Menotti RM (2011) The domestic cat as a large animal model for characterization of disease and therapeutic intervention in hereditary retinal blindness. Journal of Ophthalmology.
- Samaha G, Beatty J, Wade CM, Haase B (2019) The burmese cat as a genetic model of type 2 diabetes in humans. Anim Genet 50(4): 319- 325.
- Bergknut N, Rutges JPHJ, Kranenburg HJC, Smolders LA, Hagman R, et al. (2012) The dog as an animal model for intervertebral disc degeneration? Spine 37: 351-358.
- Struillou X, Boutigny H, Soueidan A, Layrolle P (2010) Experimental animal models in periodontology: A Review. Open Dent J 4: 37-47.
- Parng C, Seng WL, Semino C, McGrath P (2002) Zebrafish: A preclinical model for drug screening. ASSAY and drug development technologies 1(1): 41-48.
- Piato ÂL, Capiotti KM, Tamborski AR, Oses JP, Barcellos LJG, et al. (2011) Unpredictable chronic stress model in zebrafish (Daniorerio): Behavioral and physiological responses. Prog Neuropsychopharmacol Biol Psychiatry 35(2): 561-567.
- Manni I, de Latouliere L, Gurtner A, Piaggio G (2019) Transgenic animal models to visualize cancer-related cellular processes by bioluminescence imaging. Front Pharmacol.
- Martínez NFJ, Martínez MT, Mulero V, Galindo VJ (2019) Models of human psoriasis: Zebrafish the newly appointed player. Dev Comp Immunol 97: 76-87.
- Sieber S, Grossen P, Bussmann J, Campbell F, Kros A, et al. (2019) Zebrafish as a preclinical in vivo screening model for nanomedicines. Adv Drug Deliv Rev 151-152; 152-168.
- Pandey UB, Nichols CD (2011) Human disease models in Drosophila melanogaster and the Role of the fly in therapeutic drug discovery. Pharmacol Rev 63(2): 411-436.
- Gonzalez C (2013) Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat Rev Cancer 13(3): 172-183.
- Stevens CW (2011) Analgesia in amphibians: Preclinical studies and clinical applications. Vet Clin North Am Exot Anim Pract 14(1): 33-44.
- Wagner CA, Friedrich B, Setiawan I, Lang F, Bröer S (2000) The use of xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell Physiol Biochem 10(1-2): 1-12.
- Mannioui A, Zalc B (2019) Conditional demyelination and remyelination in a transgenic Xenopus laevis. Methods Mol Biol 1936: 239-248.
- Fort DJ, Mathis M (2018) Frog embryo teratogenesis assay-xenopus (FETAX): Use in alternative preclinical safety assessment. Cold Spring Harb Protoc 2018: pdb.prot098319 .
- Dias IR, Camassa JA, Bordelo JA, Babo PS, Viegas CA, et al. (2018) Preclinical and translational studies in small ruminants (Sheep and Goat) as models for osteoporosis research. Curr Osteoporos Rep 16(2): 182-197.
- McGovern JA, Griffin M, Hutmacher DW (2018) Animal models for bone tissue engineering and modelling disease. Dis Model Mech 11(4): dmm033084.
- Madsen CS, Janovitz E, Zhang R, Nguyen TV, Ryan CS, et al. (2008) The guinea pig as a preclinical model for demonstrating the efficacy and safety of statins. J Pharmacol Exp Ther 324(2): 576-586.
- Grigoleit HG, Grigoleit P (2005) Pharmacology and preclinical pharmacokinetics of peppermint oil. Phytomedicine 12(8): 612-616.
- Fureix C, Jego P, Henry S, Lansade L, Hausberger M (2012) Towards an ethological animal model of depression? A Study on Horses. PLOS ONE 7(6): e39280.
- Sapsutthipas S, Leong PK, Akesowan S, Pratanaphon R, Tan NH, et al. (2015) Effective equine immunization protocol for production of potent poly-specific antisera against calloselasma rhodostoma, Cryptelytrops albolabris and Daboia siamensis. PLoS Negl Trop Dis 9(3): e0003609.
- Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D (2010) An intrabody based on a llama single-domain antibody targeting the N-terminal α-helical multimerization domain of HIV-1 Rev prevents viral production. J Biol Chem 285(28):21768-21780.
- Pritchard JB, French JE, Davis BJ, Haseman JK (2003) The role of transgenic mouse models in carcinogen identification. Environ Health Perspect 111(4): 444-454.
- Sharpless NE, DePinho RA (2006) The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov 5(9):741-754.
- Glukhova XA, Prusakova OV, Trizna JA, Zaripov MM, Afanaseva GV, et al. (2016) Updates on the production of therapeutic antibodies using human hybridoma technique. Curr Pharm Des 22(7): 870-878.
- Gad SC (2016) Animal models in toxicology. 3rd edn, Boca Raton: CRC Press, USA, pp. 1152.
- Pinto PBA, Assis ML, Vallochi AL, Pacheco AR, Lima LM, et al. (2019) T cell responses induced by DNA vaccines based on the DENV2 E and NS1 Proteins in Mice: Importance in protection and immunodominant epitope identification. Front Immunol 10: 1522.
- Greish K, Alqahtani AA, Alotaibi AF, Abdulla AM, Bukelly AT, et al. (2019) The effect of silver nanoparticles on learning, memory and social interaction in BALB/C Mice. Int J Environ Res Public Health 16(1): 148.
- da Silva BAF, da Costa RHS, Fernandes CN, Leite LHI, Ribeiro FJ, et al. (2018) HPLC profile and antiedematogenic activity of Ximenia americana L. (Olacaceae) in mice models of skin inflammation. Food Chem Toxicol 119: 199-205.
- Borrow AP, Heck AL, Miller AM, Sheng JA, Stover SA, et al. (2019) Chronic variable stress alters hypothalamic-pituitary-adrenal axis function in the female mouse. Physiology & Behavior 209: 112613.
- Nisar S, Farooq RK, Nazir S, Alamoudi W, Alhibshi A (2019) Exposure to early life adversity alters the future behavioral response to a stressful challenge in BALB/C mice. Physiol Behav 210: 112622.
- Kim W, Hendricks GL, Lee K, Mylonakis E (2017) An update on the use of C. elegans for preclinical drug discovery: screening and identifying anti-infective drugs. Expert Opinion on Drug Discovery 12: 625-633.
- Dengg M, van Meel JCA (2004) Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. J Pharmacol Toxicol Methods 50(3): 209-214.
- Kishi N, Okano H (2017) Neuroscience research using non-human primate models and genome editing. In: Jaenisch R, Zhang F, Gage F, (Eds.) genome editing in neurosciences cham (CH): Springer, New York.
- Manickam C, Shah SV, Nohara J, Ferrari G, Reeves RK (2019) Monkeying around: Using non-human primate models to study nk cell biology in HIV Infections. Front Immunol 10: 1124.
- Pagliaccio D, Pine DS, Leibenluft E, Monte OD, Averbeck BB, et al. (2019) Cross-species convergence in pupillary response: Understanding human anxiety via non-human primate amygdala lesion. Soc Cogn Affect Neurosci 14(6): 591-599.
- Peña JC, Ho WZ (2016) Non-human primate models of tuberculosis. Microbiol Spectr 4(4).
- Tapking C, Popp D, Branski LK (2019) Pig model to test tissueengineered skin. In: Böttcher HS, Biedermann T (Eds.) skin tissue engineering: methods and protocols, Springer New York, p. 239-49.
- Dhondt L, Croubels S, Paepe PD, Cock PD, Devreese M (2019) P27 the juvenile pig as animal model for unraveling renal drug elimination processes in children. Archives of Disease in Childhood 104(6): e28-e28.
- Ferreira RN, Machado de ARA, Sanchez EF, Maria WS, Molina F, et al. (2006) Antibodies against synthetic epitopes inhibit the enzymatic activity of mutalysin II, a metalloproteinase from bushmaster snake venom. Toxicon 48(8): 1098-1103.
- Mapara M, Thomas BS, Bhat KM (2012) Rabbit as an animal model for experimental research. Dent Res J (Isfahan) 9(1): 111-118.
- Souza D, Sadananda M (2017) Anxiety and depressive-like profiles during early- and mid-adolescence in the female Wistar Kyoto rat. Int J Dev Neurosci 56: 18-26.
- Merlo E, Schereider IRG, Simões MR, Vassallo DV, Graceli JB (2019) Mercury leads to features of polycystic ovary syndrome in rats. Toxicol Lett 312: 45-54.
- Zhang RK, Wang C (2018) Effect of matrine on tumor growth and inflammatory factors and immune function in Wistar rat with breast cancer. Zhongguo Ying Yong Sheng Li Xue Za Zhi 34(4): 375-378.
- Pinheiro JEL, Boldrini FJ, de Campos ALMP, Santos NA, Bendhack LM, et al. (2018) LmrBPP9: A synthetic bradykinin-potentiating peptide from Lachesis muta rhombeata venom that inhibits the angiotensinconverting enzyme activity in vitro and reduces the blood pressure of hypertensive rats. Peptides 102: 1-7.
- Turek A, Olakowska E, Borecka A, Janeczek H, Sobota M, et al. (2016) Shape-memory terpolymer rods with 17-β-estradiol for the treatment of neurodegenerative diseases: An In Vitro and In Vivo Study. Pharm Res 33(12): 2967-2978.
- Kadhum WR, Hada T, Hijikuro I, Todo H, Sugibayashi K (2017) Development and optimization of orally and topically applied liquid crystal drug formulations. J Oleo Sci 66(9): 939-003950.
Article Type
Review Article
Publication history
Received date: June 26, 2020
Published date: July 24, 2020
Address for correspondence
Manuela Berto Pucca; Medical School of Roraima, Federal University of Roraima (UFRR), Av. Capitão Ene Garcez, Brazil
Copyright
©2020 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Oliveira IS, Alexandre-Silva GM, Cordeiro FA, Pinheiro-Júnior EL, Ferreira IG, Cerni FA, Zottich U, Pucca MB. Research Models in Biomedical Sciences: Advantages and Limitations. 2020 - 2(4) OAJBS.ID.000197.
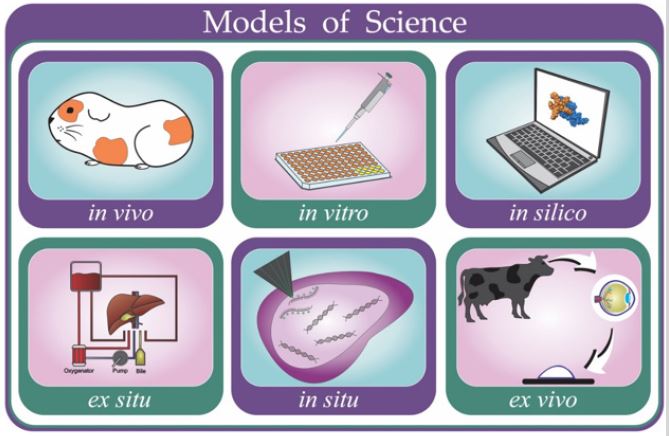
Figure 1: Research models in the biomedical sciences. In vivo - represented by a guinea pig; in vitro-represented by the 96-well plate; in silico - represented by a protein model; ex situ - represented by a perfusion system for organ preservation; in situ - represented by fluorescent in situ hybridization (FISH); and ex vivo - represented by ocular tests in bovine corneas.
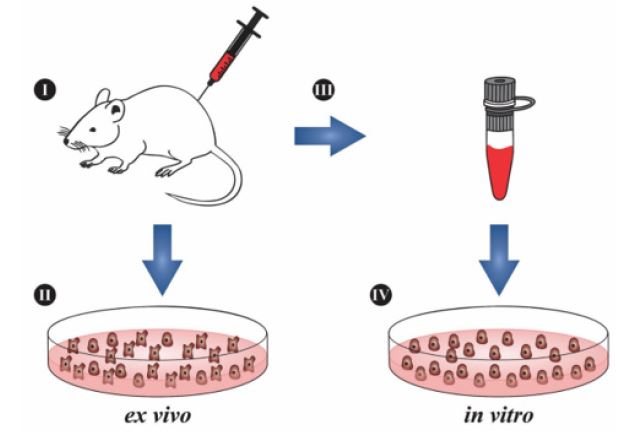
Figure 2: Difference between in vitro and ex vivo models. (I) Cells (e.g. leukocytes) are collected from an animal such as mouse; (II) differentiated cells are used right after animal collection, characterizing ex vivo assays; (III) cells taken from the animal undergo an immortalization process and are commercialized; (IV) immortalized commercial cells (cell lines) are used in assays characterizing in vitro models.
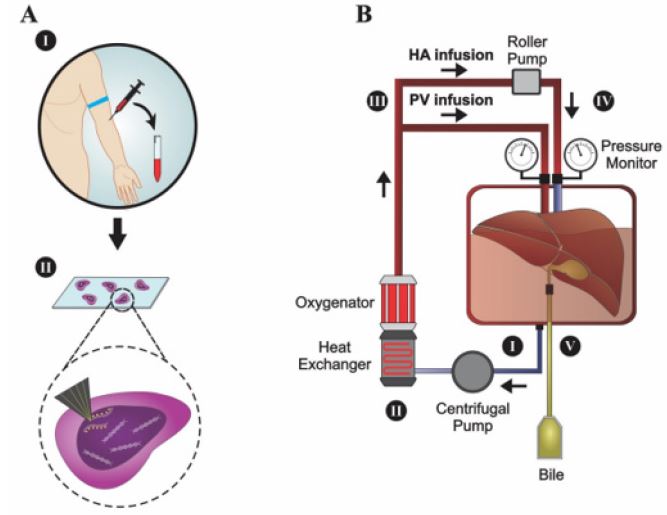
Figure 3: In situ and ex situ models. (a) Fluorescent in situ hybridization (FISH). The biological material is collected (I) and a fluorescent probe (II) is added to label the gene of interest. The material is analyzed using a fluorescence microscopic. (b) Ex situ organ perfusion. (I) The organ is placed in a container and submerged in a liquid suitable for infusions. This liquid flows to the centrifugal pump, heat exchanger and oxygenator (II). Through a bifurcation (III), the oxygenated liquid returns to the organ through the portal vein (PV) and part of it (IV) reaches a roller pump, to increase the pressure before returning to the organ through the hepatic artery (HA). (V) Container for the bile storage.
Figure 4: Main in vivo models used in preclinical research.
Table 1: In vivo models in preclinical research.