Relationship Between GDF5 +104T/C Polymorphism and the Susceptibility of Lumbar Disc Degeneration: A Meta-Analysis
ABSTRACT
Lumbar disc degeneration (LDD) is a multifactorial and chronic disease with a complex genetic background. Previous studies have reported the strong association between growth differentiation factor 5 (GDF5) +104T/C polymorphism and the risk of LDD in different ethnic populations. The aim of this study was to analyze the association between the GDF5 +104T/C polymorphism and the susceptibility of LDD by a meta-analysis. Potentially relevant studies were selected by electronic databases (PubMed, EMBASE, Web of science, the Cochrane Library Chinese National Knowledge Infrastructure) up to 10th April 2017. Summary odds ratio (OR) and 95% confidence intervals (CIs) were used to assess the strength of association between the GDF5 +104T/C polymorphism and LDD risk. Six case-control studies, including 1107 LDD cases and 4353 controls, were included. The combined results showed that GDF5 +104T/C polymorphism was significantly associated with increased susceptibility to LDD at the allele level (T vs. C: OR = 1.52, 95%CI = 1.21-1.90) and genotype level (TT vs. CC: OR = 2.43, 95%CI = 1.81-3.25; TC vs. CC: OR = 1.84, 95%CI = 1.37-2.47; TT+TC vs. CC: OR = 2.14, 95%CI = 1.61-2.84; TT vs. TC+CC: OR = 1.42, 95%CI = 1.01-1.99) in the overall population. After stratified by ethnicity, we found a strong significant association (OR = 1.80 to 3.70, all P <0.05) in Asian population and weaker associations (OR =1.20 to 1.64, all P <0.05) in Caucasian population. This meta-analysis suggests an increased risk of GDF5 +104T/C polymorphism against intervertebral disc degeneration among populations especially in Asians.
KEYWORDS
Lumbar disc degeneration; GDF5; rs143383; Polymorphism; Meta-analysis
ABBREVIATIONS:
CI: Confidence Interval; GDF5: Growth Differentiation Factor 5; LDD: Lumbar Disk Degeneration; OR: Odds Ratio; HWE: Hardy-Weinberg’s Equilibrium; SNP: Single Nucleotide Polymorphism
INTRODUCTION
Low back pain is one of the world’s most frequent disorders, and 70–85% of all people have back pain at some point in their life. It is a major source affecting quality of life and has a substantial impact on the cost of health care, leading to a significant burden of social and economic worldwide [1,2]. Lumbar disc degeneration (LDD) is the major cause of low back pain [3,4]. Although the etiology and pathogenesis of LDD are still not fully elucidated, epidemiologic studies have suggested that LDD is associated with environmental factors such as body weight, mechanical loading, physical activities and smoking [5]. In addition, heritability studies have shown that a series of genetic factors play important roles in the development of LDD [6-8]. One of the most comprehensively studied candidate genes is growth differentiation factor 5 (GDF5), a member of the transforming growth factor-β superfamily and closely correlated with bone morphogenetic proteins. Previous studies have demonstrated that GDF5 play an important role in musculoskeletal processes, affecting endochondral ossification, maintenance of tendon, formation of synovial joint and bone [9,10]. A single nucleotide polymorphism (SNP) (rs143383) involved in the regulation of GDF5 transcriptional activity has been identified in subjects carrying the T allele [11]. In the past several years, several reports have shown the association between GDF5 gene polymorphisms and the risk of osteoarthritis, congenital dysplasia of the hip. Recently, a number of research groups have reported the GDF5 +104T/C SNP is a protective factor for LDD [12-17].
A recent meta-analysis showed that there is an association between GDF5 (+104T/C) and lumbar disc degeneration in Northern European population, suggesting that GDF5 rs143383 polymorphism may be related to an elevated risk of LDD in a certain ethnic group[18]. However, these positive associations have not been analyzed in other ethnic groups. More recently, several new studies have also reported that the association of GDF5 +104T/C SNP in the promoter region might be a risk factor of LDD in Asian population.
Therefore, this study aimed to summarize the current data and update the meta-analysis, which may minimize potential publications bias, enlarge the sample size and provide more solid evidence. In the present study, a meta-analysis was performed to assess the associations between GDF5 +104T/C polymorphism and the susceptibility of the LDD in different ethnic populations.
RESULTS
Study Characteristics
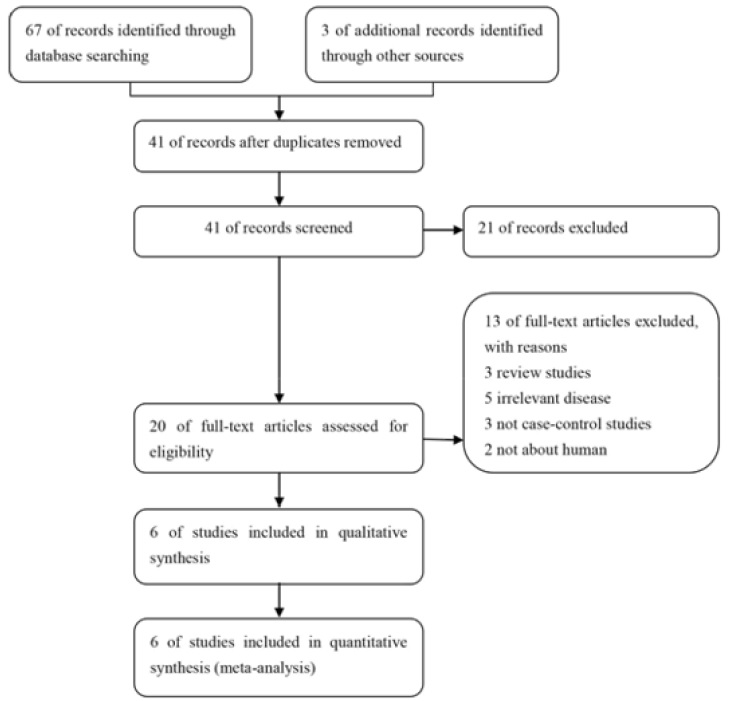
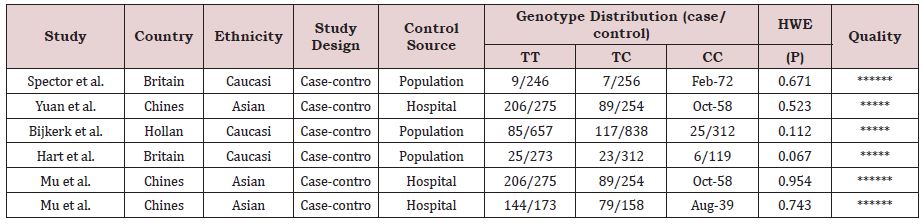
As shown in Figure 1, a total of 270 studies were obtained from databases after initial search, and three of them were selected from the reference lists of the identified publications. After first screening, 79 of them were excluded based on the selection criteria. And 172 of records were excluded for improper title/ abstract. Among the remaining 19 studies, 3 were review studies; 5 reported other disease; 3 were not case-control studies and 2 were not about human. Finally, 6 eligible studies were included in this meta-analysis, including 1107 cases and 4353 controls. The detailed characteristics of the included articles are shown in Table 1. Genotype distributions among the controls of all included studies were corresponded to HWE. The 6 case–control studies were published between 1994 and 2014. Among them, 3 studies were conducted in Caucasian and 3 in Asian.
Meta-Analysis Results
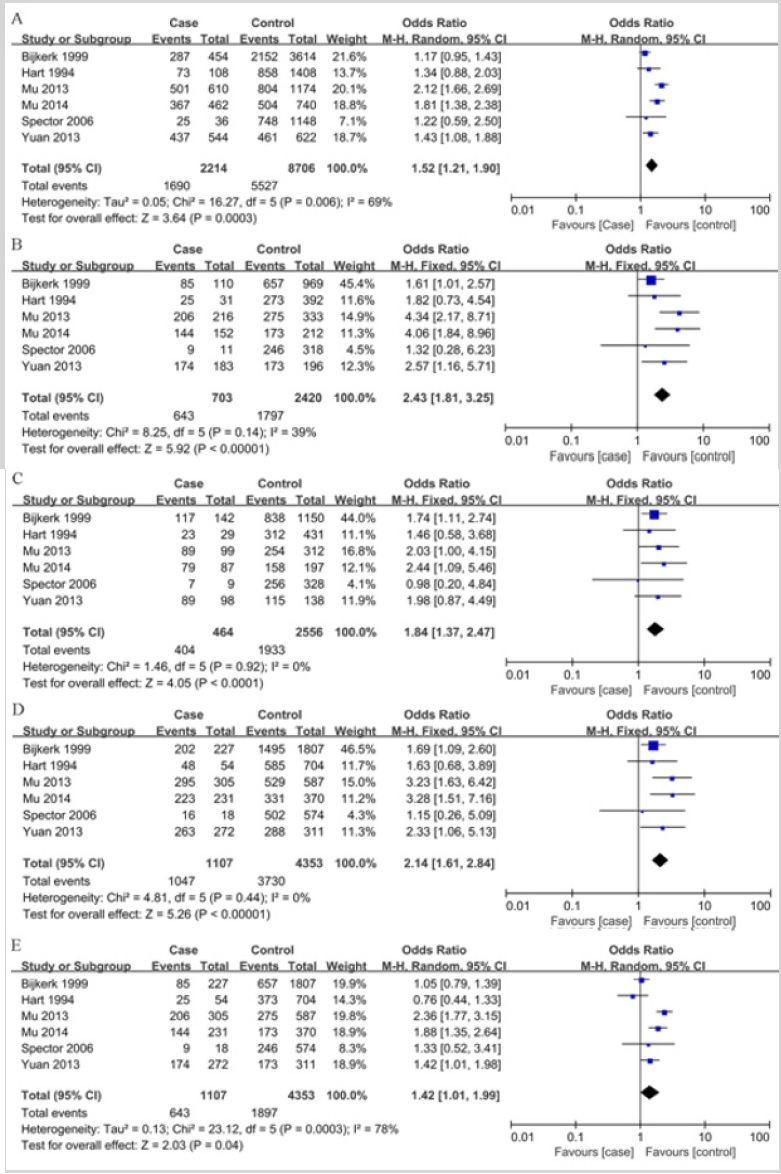
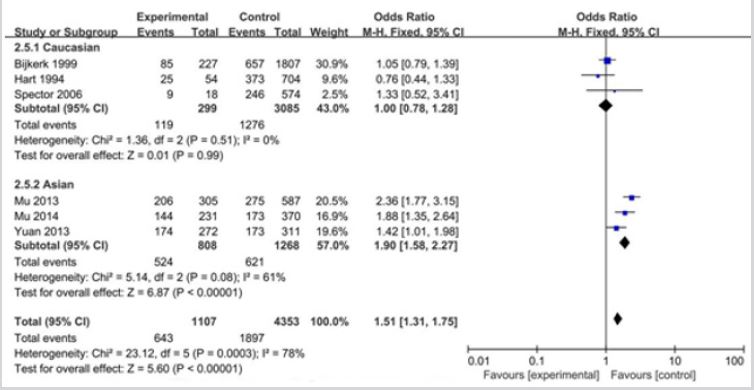
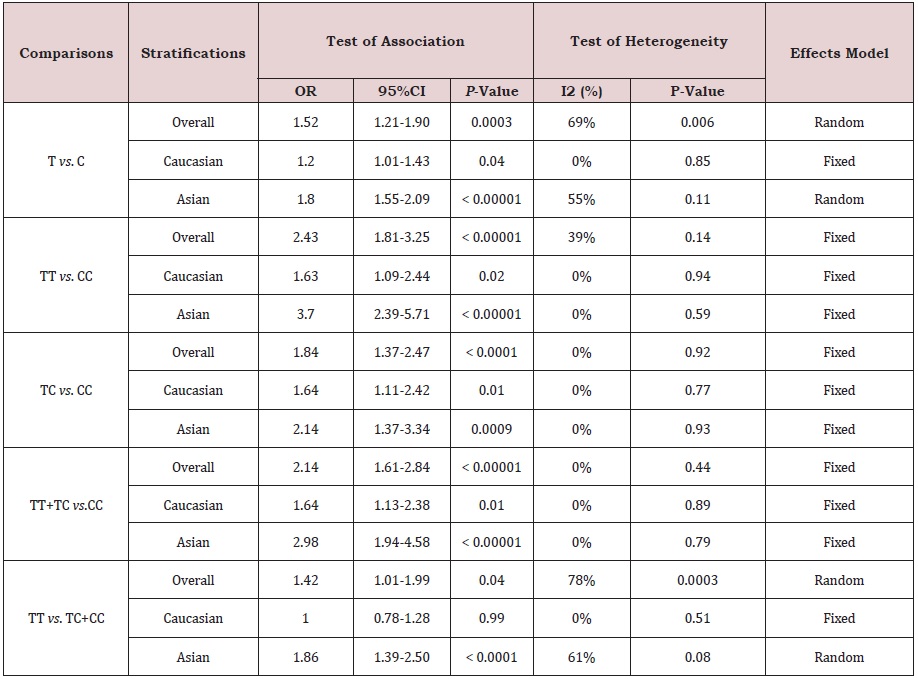
As shown in Table 2 and Figure 2, The association was found between the GDF5 +104T/C polymorphism and LDD susceptibility in all the genetic models: T vs. C (OR = 1.52, 95%CI = 1.21-1.90, P<0.001), TT vs. CC (OR = 2.43, 95%CI = 1.81-3.25; P<0.001), TC vs. CC (OR = 1.84, 95%CI = 1.37-2.47, P<0.001), TT+TC vs. CC (OR = 2.14, 95%CI = 1.61-2.84, P<0.001), TT vs. TC+CC (OR = 1.42, 95%CI = 1.01-1.99, P = 0.04) (Figure 2). In the subgroup analysis stratified by ethnicity, studies were divided into Caucasian and Asian populations. The same association was found in the Caucasian (T vs. C: OR = 1.20, 95%CI = 1.01-1.43, P = 0.04; TT vs. CC: OR = 1.63, 95%CI = 1.09-2.44; P = 0.02; TC vs. CC: OR = 1.64, 95%CI = 1.11-2.42, P = 0.01; TT+TC vs. CC: OR = 1.64, 95%CI = 1.61-2.84, P = 0.01) and Asians (T vs. C: OR = 1.80, 95%CI = 1.55-2.09, P<0.001; TT vs. CC: OR = 3.70, 95%CI = 2.39-5.71; P<0.001; TC vs. CC: OR = 2.14, 95%CI = 1.37-3.34, P<0.001; TT+TC vs. CC: OR = 2.98, 95%CI = 1.94-4.58, P<0.001; TT vs. TC+CC: OR = 1.86, 95%CI = 1.39-2.50, P<0.001) groups. But the GDF5 +104T/C polymorphism was not significantly associated with LDD in Caucasians when comparing TT vs. TC+CC genotypes (OR = 1.00, 95 % CI = 0.78-1.28, P = 0.99). (Figure 3); (Table. 2).
Heterogeneity Analysis
In Q test, significant heterogeneity was detected in allele T vs. allele C model (P=0.006) and TT vs. TC+CC model (P=0.0003). Therefore, the random-effects model was used to evaluate overall results in these two models, while the fixed-effects model was selected for the other three models (TT vs. CC: P = 0.14; TC vs. CC: P=0.92; TT+TC vs. CC: P=0.44). Notably, the significant heterogeneity of that two models was considerably decreased after subgroup analysis by ethnicity, which might contribute to the heterogeneity.
Sensitivity Analysis
We removed one single study from the overall pooled analysis each time and recalculated overall outcomes to evaluate the impact of each included study on pooled results. The pooled ORs were not substantially changed, suggesting that our results were statistically stable and reliable.
Publication Bias
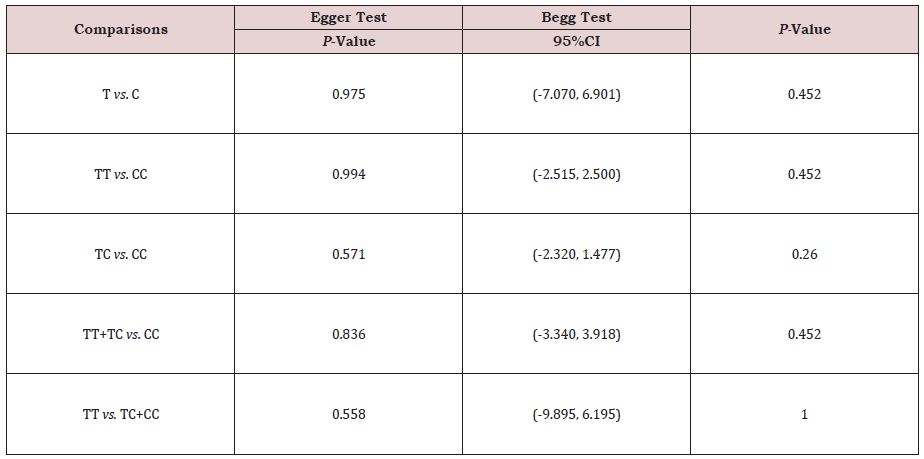
Both Begg’s test and Egger’s test were used to evaluate the possibility of existing significant publication bias. No significant publication bias was found in the Begg’s test and Egger’s test (P>0.05) (Table 3).
DISCUSSION
The process of LDD is multifactorial and chronic including intrinsic and extrinsic determinants. A number of environmental factors may contribute to etiology and pathological of disc degeneration, such as mechanical overloading, age, obesity, smoking status, but it is still far from being clear at present [19- 21]. Recently, many polymorphisms in genes have been shown to be associated with LDD, which will bring positive significance for the prevention and treatment of LDD [22,23]. Previous studies have reported the GDF5 genetic polymorphisms play a critical role in etiology and pathogenesis of disc degeneration [18]. The association between GDF5 gene polymorphism and LDD susceptibility has been validated in large-sample epidemiological studies with diverse ethnic backgrounds. In the present study, a meta-analysis was designed in order to update and investigate the correlation between the GDF5 +104T/C polymorphism and the susceptibility to LDD in different ethnic populations. Six studies related to GDF5 +104T/C polymorphism were included with a total of 1107 LDD patients and 4353 controls in the present metaanalysis. It revealed significant evidence of association between the GDF5 +104T/C polymorphism and LDD. These results indicate that T allele of GDF5 was significantly related to a higher risk for LDD in Caucasian and Asian subjects. In the subgroup analysis by ethnicity, GDF5 +104T/C polymorphism was still found to be significantly related to LDD in Caucasian and Asian subjects, although not significantly related to LDD in Caucasians when evaluated using a dominant model (TT vs. TC+CC: OR = 1.00, 95 % CI = 0.78-1.28, P = 0.99). Moreover, effects sizes in Asian populations were detected to be consistently greater than in Caucasian populations, suggesting that this gene polymorphism may play different roles in LDD susceptibility among different ethnic origins.
Heterogeneity is a potential problem to be considered when comprehending the results of meta-analyses. In this meta-analysis, significant heterogeneity between different studies existed in the overall population when using an allele model (T vs. C) and a recessive model (TT vs. TC+CC). After subgroups analysis by ethnicity, the heterogeneity had attenuated or disappeared in populations of Caucasian or Asian descent. That means ethnicities with different genetic background might contribute to the heterogeneity. Moreover, we carried out sensitivity analyses for GDF5 +104T/C. Removal one single study from the overall pooled analysis each time do not impact on overall outcomes, suggesting the reliability and stability of the results.
Some limitations of this meta-analysis should be acknowledged. First, the number of published studies included was small for a comprehensive analysis. Some unpublished, non-English, conference articles or studies without sufficient original data were not included in our meta-analysis, which may lead to a bias of our results. Second, we failed to conduct stratification analysis by age, sex, smoking status, obesity, physical load, or other lifestyle factors because of the data limitation of the included studies. Third, other polymorphic loci and environmental factors have been associated with LDD. We could not address gene-gene or gene-environmental interactions in this meta-analysis due to the lack of relevant information. Therefore, additional studies with larger sample sizes of lumbar disc degeneration and controls are required.
In summary, the current meta-analysis comprehensively suggests that the GDF5 +104T/C polymorphism is significantly associated with risk of LDD susceptibility, particularly in Asians. Because of the limitations of this study, future research on GDF5 +104T/C polymorphisms and LDD susceptibility are necessary and gene-gene and/or gene-environmental interactions of other genetic and environmental factors should be investigated.
MATERIALS AND METHODS
Literature Searching Strategy
Potentially relevant studies were selected by PubMed, EMBASE, Web of science, the Cochrane Library Chinese National Knowledge Infrastructure (CNKI) databases up to 10th April 2017. The following keywords were used: (“GDF5”or “growth differentiation factor 5” or“rs143383” or “+104T/C”) and (“polymorphism” or “SNPs” or “single nucleotide polymorphisms”) and (“disc degeneration” or “disc” or “disc herniation” or “intervertebral disc degeneration” or “low back pain”). In addition, manually screening of the references in selected articles were performed for other potentially relevant papers. All research was limited to English and Chinese.
Inclusion and Exclusion Criteria
The studies meeting the following inclusion criteria were included: (1) the study should evaluate the relationship between GDF5 gene +104T/C polymorphism and susceptibility of the LDD; (2) independent case-control studies for humans; (3) sufficient data on genotype distributions in both cases and controls for calculating an odds ratio (OR) with 95% confidence interval. We excluded reviews, case reports, abstracts or animal studies. If studies were duplicated or overlapped publications, the most recent publications were included. All identified studies were selected by two investigators independently.
Data Extraction
According to the inclusion and exclusion criteria, two investigators (Liang and Deng) reviewed and extracted data independently. The following information was extracted from each study: the first author’s name, year of publication, country, ethnicity, source of control; study design, sample size, allele numbers and genotype frequencies in cases and controls. When any inconsistencies appeared, we resolved it through discussion. If not, a third reviewer was invited to make a final decision.
Statistical Analysis
The Hardy-Weinberg equilibrium (HWE) in controls for each study was assessed by a goodness of fit chi-square test before statistical analysis. Pooled ORs with 95% CIs were calculated to estimate the strength of association between GDF5+104T/C polymorphism and susceptibility of LDD under five models: TT vs. CC, TT+TC vs. CC, TT vs. CC+TC, allele T vs. allele C and TC vs. CC. P<0.05 was considered as statistically significant. Z-test was used to evaluate the significance of the pooled OR. I2-statistics and Q-test were used to evaluate the statistical heterogeneity among studies, which was considered as significant when PQ<0.10 or I2>50% [24]. Then, the overall or pooled OR was obtained by a random-effect (DerSimonian-Laird method) in the presence of heterogeneity (PQ<0.10 or I2>50%).
Otherwise, a fixed-effect model (Mantel- Haenszel method) [liwen3-18] was selected (PQ>0.05 or I2<50%) [25]. To further test that relationship and explore the sources of heterogeneity, we performed subgroup analysis based on ethnicity. Sensitivity analyses were performed to assess the influence of each single study on pooled results. We used Begg’s funnel plot and Egger’s test to measure potential publication bias [26,27]. All statistical analyses were performed using software RevMan 5.3 (The Cochrane Collaboration) and STATA 12.0 (StataCorp, College Station, TX).
CONCLUSION
There is a strong significant association (OR = 1.80 to 3.70, all P<0.05) in Asian population and weaker associations (OR =1.20 to 1.64, all P<0.05) in Caucasian population. This meta-analysis suggests an increased risk of GDF5 +104T/C polymorphism against intervertebral disc degeneration among populations especially in Asians.
ACKNOWLEDGEMENT
Thanks for the support of Liang to Deng as the Senior, you help me complete this paper as well as the challenge of life, I hope to work with you all the time.
FUNDING
This study was supported by grants 2016YFC1100100 from The National Key Research and Development Program of China and by grants 91649204 from Major Research Plan of National Natural Science Foundation of China.
AUTHOR CONTRIBUTIONS
Liang contributed to the idea of the current study. Liang and Deng made a search of articles and screened them independently. Any conflict was solved by consulting the senior author (Shao). Deng and Liang extracted data from the six final studies and make tables. Liang and Chen played an important function in analyzing the results and the data analyses and make pictures. Shao polished the draft and approved the final version.
REFERENCES
- Andersson GB (1999) Epidemiological features of chronic low-back pain. Lancet 354(9178): 581-585.
- Katz JN (2006) Lumbar disc disorders and low back pain: socioeconomic factors and consequences. J Bone Joint Surg Am 88 Suppl 2: 21-24.
- Luoma K, Riihimaki H, Luukkonen R, Raininko R, Viikari Juntura E, et al. (2000) Low back pain in relation to lumbar disc degeneration. Spine (Phila Pa 1976) 25(4): 487-492.
- Borenstein DG (2001) Epidemiology, etiology, diagnostic evaluation, and treatment of low back pain. Curr Opin Rheumatol 13(2): 128-134.
- Battie MC, Videman T, Levalahti E, Gill K, Kaprio J, et al. (2008) Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine (Phila Pa 1976) 33(25): 2801-2808.
- Battie MC, Videman T (2006) Lumbar disc degeneration: Epidemiology and genetics. J Bone Joint Surg Am 88 Suppl 2: 3-9.
- Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, et al. (2013) Genetic polymorphisms associated with intervertebral disc degeneration. Spine J 13(3): 299-317.
- Martirosyan NL, Patel AA, Carotenuto A, Kalani MY, Belykh E, et al. (2016) Genetic Alterations in Intervertebral Disc Disease. Front Surg 3: 59.
- Nishitoh H, Ichijo H, Kimura M, Matsumoto T, Makishima F, et al. (1996) Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor- 5. J Biol Chem 271(35): 21345-21352.
- Mikic B, Battaglia TC, Taylor EA, Clark RT (2002) The effect of growth/ differentiation factor-5 deficiency on femoral composition and mechanical behavior in mice. Bone 30(5): 733-737.
- Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, et al. (2007) A functional polymorphism in the 5’ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 39(4): 529-533.
- Yuan K FJ, Cui ZM, Zhao JH, Dong QR (2013) Association between single nucleotide polymorphism of growth differentiate factor 5 and lumbar disc degeneration. Jiangsu Med J 39(18): 2147-2149.
- Mu J, Ge W, Zuo X, Chen Y, Huang C, et al. (2013) Analysis of association between IL-1β, CASP-9, and GDF5 variants and low-back pain in Chinese male soldier: clinical article. J Neurosurg Spine 19(2): 243-247.
- Mu J, Ge W, Zuo X, Chen Y, Huang C, et al. (2014) A SNP in the 5’UTR of GDF5 is associated with susceptibility to symptomatic lumbar disc herniation in the Chinese Han population. Eur Spine J 23(3): 498-503.
- Hart DJ, Mootoosamy I, Doyle DV, Spector TD (1994) The relationship between osteoarthritis and osteoporosis in the general population: the Chingford Study. Ann Rheum Dis 53(3): 158-162.
- Bijkerk C, Houwing Duistermaat JJ, Valkenburg HA, Meulenbelt I, Hofman A, et al. (1999) Heritabilities of radiologic osteoarthritis in peripheral joints and of disc degeneration of the spine. Arthritis Rheum 42(8): 1729-1735.
- Spector TD, Williams FM (2006) The UK adult twin registry (TwinsUK). Twin Res Hum Genet 9(6): 899-906.
- Williams FM, Popham M, Hart DJ, De Schepper E, Bierma Zeinstra S, et al. (2011) GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in Northern European women. Arthritis Rheum 63(3): 708-712.
- De Schepper EI, Damen J, Van Meurs JB, Ginai AZ, Popham M, et al. (2010) The association between lumbar disc degeneration and low back pain: the influence of age, gender, and individual radiographic features. Spine (Phila Pa 1976) 35(5): 531-536.
- Samartzis D, Karppinen J, Mok F, Fong DY, Luk KD, et al. (2011) A population-based study of juvenile disc degeneration and its association with overweight and obesity, low back pain and diminished functional status. J Bone Joint Surg Am 93(7): 662-670.
- Dario AB, Ferreira ML, Refshauge KM, Lima TS, Ordonana JR, et al. (2015) The relationship between obesity, low back pain and lumbar disc degeneration when genetics and the environment are considered: a systematic review of twin studies. Spine J 15(5): 1106-1117.
- Kalichman L, Hunter DJ (2008) The genetics of intervertebral disc degeneration. Associated genes. Joint Bone Spine 75(4): 388-396.
- Hanaei S, Abdollahzade S, Khoshnevisan A, Kepler CK, Rezaei N, et al. (2015) Genetic aspects of intervertebral disc degeneration. Rev Neurosci 26(5): 581-606.
- Attia J, Thakkinstian A, D Este C (2003) Meta-analyses of molecular association studies: Methodologic lessons for genetic epidemiology. J Clin Epidemiol 56(4): 297-303.
- Der Simonian R (1996) Meta-analysis in the design and monitoring of clinical trials. Stat Med 15(12): 1237-1248.
- Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50(4): 1088-1101.
- Egger M, Davey Smith G, Schneider M, Minder C, et al. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109): 629- 634.
Article Type
Research Article
Publication history
Received date: October 21, 2020
Published date: October 29, 2020
Address for correspondence
Xiangyu Deng, Department of Orthopaedic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, People’s Republic of China
Copyright
©2020 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Xiangyu D, Hang L, Zengwu S. Relationship Between GDF5 +104T/C Polymorphism and the Susceptibility of Lumbar Disc Degeneration: A Meta-Analysis. 2020 - 2(5) OAJBS.ID.000231.
Figure 1: Flow diagram of the study selection process.
Figure 2: Forest plots for association between GDF5 +104T/C polymorphism and LDD risk in different genetic models. ((A) Allele model (T vs. C); (B) Homozygote model (TT vs. CC); (C) Heterozygote model (TC vs. CC); (D) Dominant mode (TT+TC vs. CC); (E) Recessive model (TT vs. TC+CC)).
Figure 3: Forest plots for association between GDF5 +104T/C polymorphism and LDD risk based on ethnicity. ((A) Allele model (T vs. C); (B) Homozygote model (TT vs. CC); (C) Heterozygote model (TC vs. CC); (D) Dominant model (TT+TC vs. CC); (E) Recessive model (TT vs. TC+CC)).
Table 1: Characteristics of studies included in the meta-analysis.
Table 2: Meta-analysis of the association of GDF5 (+104T/C) polymorphism with LDD risk.
Table 3: Publication bias tests for association between GDF5 +104T/C polymorphism and LDD.