In Silico TERT-Gene Knocked Out Via CRISPR/Cas9 for Studying the Possibility of Curing Cancerous Cells
ABSTRACT
Due to the growing spread of cancer in different countries, as well as global attention to CRISPR/cas9 gene-editing system, we
decided to suggest a method for treating a variety of cancers by CRISPR/cas9 through the use of in silico analysis and information
available on online soft wares. In order to achieve the goal of cancer treatment, telomerase gene (TERT-gene) was selected as the
target gene for its prevalence of 80-90% in cancers. Telomerase is silent and inactive in most somatic cells. However, this gene is
active in the majority of tumors and cancer cells. Activation of this gene leads to the immortality of cancer cells, therefore knocking
out the telomerase gene by CRISPR/cas9, which has the ability to precisely cut and edit genome can provide a method to treat a
variety of cancers. In order to in silico design the appropriate gRNAs to silence the telomerase gene we used data available on NCBI
website (www.ncbi.nlm.nih.gov) and the evaluation and anticipation of cutting PAMs through CRISPOR website (www.crispor.tefor.
net). In this regard two gRNAs, one for 5’ and the other for 3’ site of promoter of telomerase gene were designed, to remove the
target promoter and silence the desired gene. Therefore, in separate stages, 180bp sequences from the beginning and the end of the
promoter of telomerase gene were placed on the crispor website and probable PAMs in these two regions were evaluated according
to efficiency and off targets, to finally select and design the most appropriate PAM and following to its gRNA. In the 180bp region
of 5’ site of telomerase gene promoter, 9 PAMs in the correct direction (reverse) were found among which the PAM in the position
of “81rev” got the highest score regarding to the most efficiency and specificity and the least off targets. In the 180 bp region of 3’
site of telomerase gene promoter, 8 PAMs in correct direction (forward) were found, however, unfortunately none of them got the
adequate score due to excess of GC content and also low specificity. Therefore, a region of 180 bp from downstream of promoter was
selected. This time 8 PAMs in the correct direction (forward) were found from which a PAM in the position of “159 fw” got the best
score. According to selected PAMs, the proposed gRNAs are as followed:
5’ gRNA: CAT GGC GAG GAA ACG CCT CC CGG
3’ gRNA: GCT GCG CAG CCA CTA CCG CG AGG
KEYWORDS
Cancer: CRISPOR: CRISPR/cas9: Gene knocked out: NCBI: TERT gene
INTRODUCTION
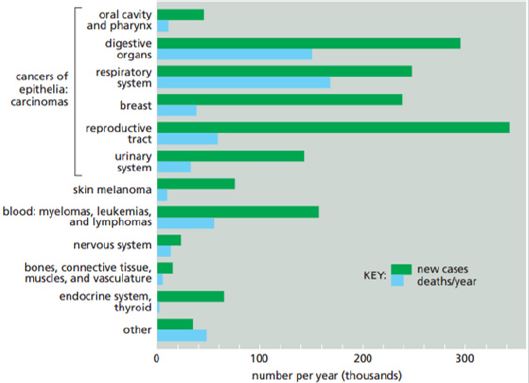
Cancer is one of the most common causes of death in many societies today (Figure 1). The disease occurs due to a variety of genetic and environmental conditions that cause an uncontrolled growth of cells. Some of the features and general differences between cancerous cells and ordinary ones are:
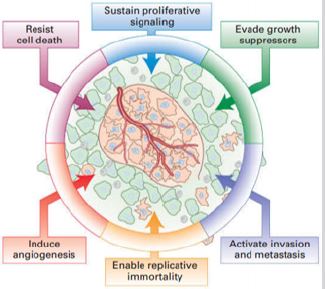
a) They continue proliferating in the absence of growth
factors or receiving growth signals
b) Growth inhibitors do not have any effect on them
c) They become resistant to apoptosis
d) When a group of fast-growing cancer cells are gathered as
tumor, they start angiogenesis for better nourishment of all cells
e) They become immortal
f) In the worst condition some of the cancerous cells are
separated and start metastasis, though start spreading to other
parts of the body.
Genetic changes and aneuploidy also occur in different cancers, but these changes are different among people and cancers [1]; (Figure 2). The multiple hit model express that a cell should undergo several mutations to become cancerous. For instance, activation of one oncogenic gene by itself will not lead to cancer, for about 5 to 6 hits or mutations are needed in order to push an ordinary cell into a cancerous one [1]. According to last findings, about 1 to 10 mutations are needed to create cancer, which also depends on the cancer type [2]. Therefore, considering that in the following study we pursuit genome editing, we look for a special mutation or an inclusive gene to not only treat cancer but also be used for an extensive range of cancers.
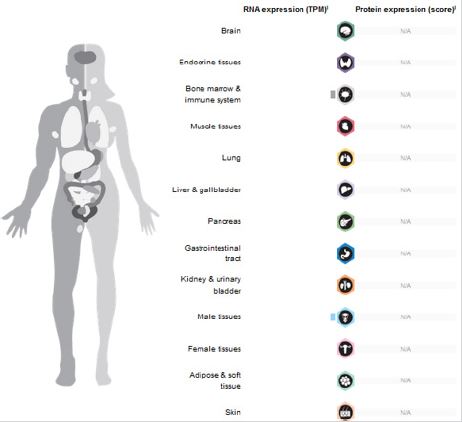
There is a GT rich region at the end of linear chromosomes called telomere. Telomerase enzyme is responsible for the maintenance and protection of this terminal region from shortening. In 1970 it was found that DNA polymerase enzyme is not capable of transcribing the terminal region of telomeres (End replication problem phenomena), and that many of human cells, except of stem cells have deficiencies in telomerase, which would lead to cell senescence [3,4]. In most cancer cells the telomerase gene is activated and will provide the immortality of cancer cells [5-7]. Even in some cancers telomere is maintained and protected by some mechanisms without telomerase activity that are called Alternative Lengthening of Telomerase (ALT). Before birth telomerase becomes inactive in most somatic cells so that except of stem cells, prevent the unstoppable proliferation of cells [8]; (Figure 3). It is remarkable that some studies have shown that telomerase gene are active in cancerous stem cells [9-11].
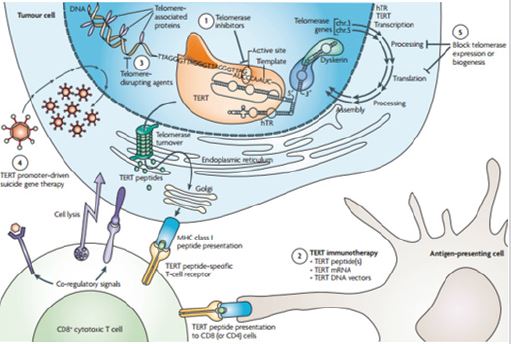
Therefore, efforts for knocking out telomerase gene or removing its protein is an action which may be able to treat a broad range of cancers, because in about 80-90% of cancers, telomerase is active and almost no other gene has this amount of extension in cancers [11,12]. So far different therapeutic approaches have been used to remove telomerase and cure cancer, some of them are:
a) Enzyme suppressor
b) Telomerase immunotherapy, in which the immune system
is stimulated to attack the telomerase-active cells.
c) Manipulation of telomerase enzyme in order to block the
ability of matching or recognizing the telomeres.
d) In this method regarding that the cancerous cell activates
the promoter of telomerase gene, the promoter is placed in the upstream region of a gene whose expression leads to a product
inducing death in cancer cells.
e) Blocking the expression or function of telomerase
pathway [11]; (Figure 4).
According to last findings, whereas the frequency of TERT gene is high in many cancers, but the accurate molecular mechanism which make this phenomenon is not clear until now. Somatic mutations in coding region of TERT gene in human tumors are rare, while somatic and germ line mutations in promoter of this gene in melanoma and human cancerous cell lines are very prevalent. These mutations mostly occur in two region “-124” and “-146” bp upstream of the “ATG” start point. In the following table, the frequency of promoter mutations in some kind of cancers have been showed (Table 1):
According to mentioned points the importance of silencing telomerase gene to cure many tumors and cancers is notable. Clearly removing the promoter of telomerase gene can highly treat cancer and cause telomerase silencing and inactivation. So, regarding a fast, accurate and low cost gene editing system we discuss CRISPR genome editing tool.
Three common genome editing tools are: ZFN (Zinc Finger Nucleases), TALENS (Transcription Activator-Like Effector Nuclease), and CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat). ZFN and TALENZ are chimeric nucleases with restriction enzyme Fok1, in which the nonspecific cutting protein part attaches the specific attaching enzyme. But CRISPR/ cas9 system, which was first found in Streptococcus Pyogenes as an immune system, consist of two RNA regions called crRNA and tracer RNA, which are complement to each other in some parts. Some parts of crRNA are hybridized to target DNA and recognize it. The duty and role of tracer RNA is absorption of cas9 protein to RNA complex. In synthetic experiments both RNAs are designed as one RNA called gRNA [13,14]. So, in designing CRISPR system unlike other editing systems there is no need of protein manipulation but just designing an appropriate gRNA can have different applications. So, working with CRISPR is easier and cheaper. In addition, PAM 3-nucleotide sequence (protospacer adjacent motif) which exist in the structure of crRNA and helps to better and more accurately recognize and cleave the target sequence. Therefore, considering accuracy CRISPR system has a higher capacity compared to others [13]. Gene deletion is one of the most commonly used cases by CRISPR, for it is easily detected by PCR. It can often produce a loss of function mutation and can be helpful for the study of noncoding factors [15]. However, CRISPR is used in different purposes of knocking in or knocking out genes [16,17], genome library screening [18,19], and variable other cases. Therefore, CRISPR system is highly flexible that has the ability of long deletions [20], and even epigenetic studies [14, 21]. Sometimes by manipulating cas9 protein we can inactive its endonuclease activity to use it for other purposes [22,23]. Using this system for the treatment of genetic disorders is improving, from which we can consider curing muscular dystrophy and Huntington models [24,25].
OBJECTIVES
Given the growing importance of cancer treatment, it is important to find a precise, fast and inexpensive method, and a way which respond to more types of cancers. Therefore, in this article telomerase gene has been selected as the target gene, active in 80- 90% of cancers, to be knocked out and disabled by the CRISPR/ cas9 gene-editing system which is highly accurate. Because CRISPR system only requires designing of gRNA, and unlike some geneediting methods does not need protein design and manipulation, the designs and costs are relatively appropriate.
MATERIALS AND METHODS
First on NCBI website (www.ncbi.nlm.nih.gov) the sequence of the promoter and a part of cdc of TERT gene was identified (Figure 5). Then, considering the fact that for removing a sequence we need two gRNAs, one at 5’ end and the other at 3’ end, so one 180bp length sequence at the beginning of promoter gene and the other with the same length (180 bp) from the end of the gene were selected. The promoter of telomerase gene has a length of 1665bp.
The query sequence at the 5’ end of TERT-gene promoter (180bp):
atcatcagct tttcaaagac acactaactg cacccataat actggggtgt cttctgggta tcagcgatct tcattgaatg ccgggaggcg tttcctcgcc atgcacatgg tgttaattac tccagcataa tcttctgctt ccatttcttc tcttccctct tttaaaattg tgttttctat
The query sequence at the 3’ end of TERT-gene promoter (180bp):
cggag ggactgggga cccgggcacc cgtcctgccc cttcaccttc cagctccgcc tcctccgcgc ggaccccgcc ccgtcccgac ccctcccggg tccccggccc agccccctcc gggccctccc agcccctccc cttcctttcc gcggccccgc cctctcctcg cggcgcgagt ttcag
The query sequence at the downstream of TERT-gene promoter (180bp):
cttcctttcc gcggccccgc cctctcctcg cggcgcgagt ttcaggcagc gctgcgtcct gctgcgcacg tgggaagccc tggccccggc cacccccgcg atgccgcgcg ctccccgctg ccgagccgtg cgctccctgc tgcgcagcca ctaccgcgag gtgctgccgc tggccacgtt
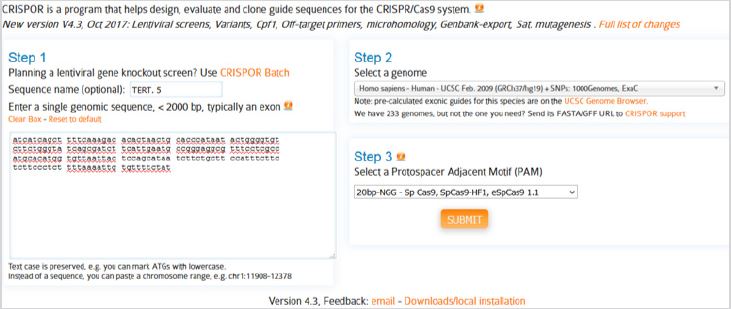
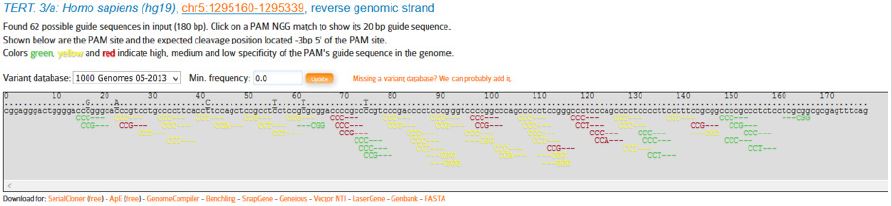
In the next stage CRISPOR website (www.crispor.tefor.net) was opened and for the first step we submit the selected sequence. In the second and third steps, which are human genome and types of selected PAMs respectively, we use software defaults (Figure 6). After submitting the sequence, available PAMs in the sequence, according to the types of selected PAMs, NGG, are presented. In the 5’ region of telomerase gene promoter 18 PAMs were found from which 9 of them had correct direction (reverse) and 9 of them had opposite direction (forward).
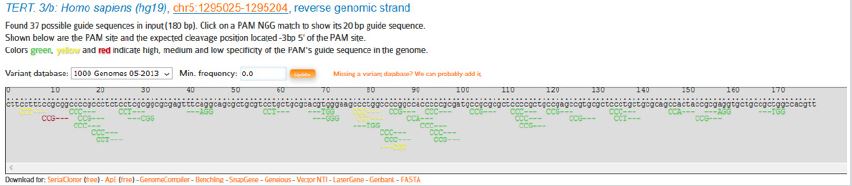
The same procedure was used for the 180bp-sequence of the 3’ end of promoter. This sequence included 62 PAMs, 8 of which had the correct direction (forward) and just 2 of these PAMs had high specificity; however, both of them had high CG content. According to the fact that high GC content (>50%) in gRNA is inappropriate due to the tight binding with target sequence, none of PAMs in this region were appropriate. Therefore, some of the downstream sequences of the promoter, including some of the first cdc and mRNA, were added to the removal region to get the appropriate PAM in order to be able to cut and eliminate telomerase gene promoter. So 180 bp region downstream of the promoter, which 45bp is related to the promoter, and the other 135 bp is related to mRNA, of which 80 bp is also part of the first cdc, were selected to find the desired PAM. Same as previous sequences, this region was also submitted on CRISPOR website and the result was 37 found PAMs from which 8 PAMs had the correct direction (forward) and the rest were in the opposite direction.
RESULTS
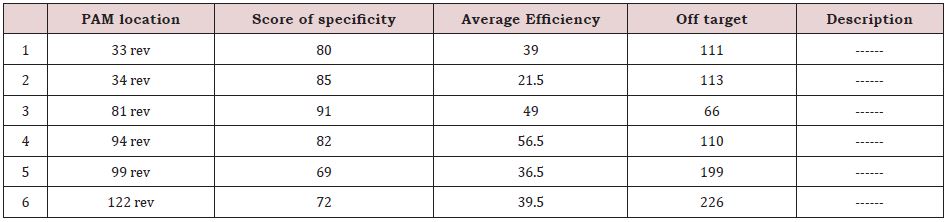
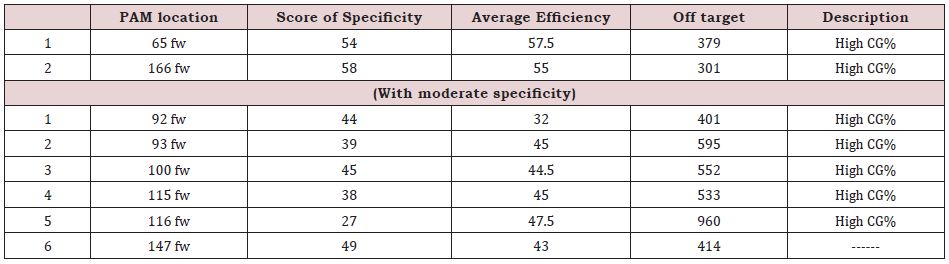
As it was mentioned, in the 5’ region, 9 PAMs (Figure 7) were in the desired direction (reverse), among which only PAMs with high specificity were analyzed and evaluated as follows (Table 2):
According to the scores in the above table, the PAM in the “81 rev” position was selected for the highest specificity, and efficiency as well as the lowest off target. The 20 bp gRNA sequence regarding this PAM is as follows: 81 rev gRNA: CAT GGC GAG GAA ACG CCT CC CGC
In the case of the 180 bp sequence of the 3’ end of telomerase gene promoter, from 8 PAMs in forward direction (Figure 8), 2 of them had high specificity and the other 6 ones had moderate specificity and the scores are as follows (Table 3):
According to above table, due to high CG content and high off targets, none of the PAMs were appropriate. Therefore, in the following table, 180bp sequence of promoter’s downstream was evaluated (Figure 9).
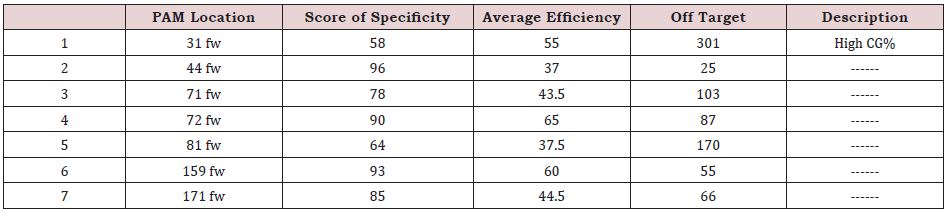
This region had 8 PAMs in the right direction, which 7 of them have high specificity (Table 4). According to this table, PAMs in “44 fw” and “159 fw” had the best conditions, so to find the best PAM, we examined the two gRNAs of mentioned PAMs. 44 fw gRNA: CTC CTC GCG GCG CGA GTT TC AGG 159 fw gRNA: GCT GCG CAG CCA CTA CCG CG AGG
Regarding the two sequences above, the gRNA of “159 fw” PAM was selected due to its higher CG content compared to “44 fw” because it helps better binding. It is noteworthy that among the 7 PAMs of this region, only “31 fw” position had an inappropriate high CG content.
DISCUSSION
Using in silico methods for the purpose of pre-designing the gRNAs in order to cleave and eliminate a gene or any kind of gene expression process by CRISPR/cas9 system is important because it is possible to examine PAMs and the probable locus in the target gene area and it also provides the researcher with the selection of the most appropriate and most specific PAM. This will result in finding the best gRNA, reducing costs, increasing work accuracy and reducing work time. However in vitro experiments are needed to investigate and prove the efficiency of the designed gRNAs, lack of toxicity or having any trouble. And following to it in vivo method and clinical experiments are necessary for final approvals of this study. A remarkable point in clinical stages is to look at the off targets and their side effects. To do this complete genomic sequencing can be effective in detecting possible mutations resulting from the use of CRISPR/cas9 editing system. Provided that sequencing should be performed prior to manipulation, so that by evaluating the mutations before and after using CRISPR, the side effects of this approach can be achieved [26,27].
CONCLUSION
The type of lifestyle in modern societies has caused a high incidence of cancers. Given the fact that cancer is a multifactorial disease (multiple hit theory) and requires several genetic abnormalities for its occurrence, it is difficult to find a single method for the treatment of all cancers. One of the similar factors in 80-90% of all types of cancers is activation of telomerase gene and immortality of cancer cells. Therefore, finding and designing appropriate gRNAs, to remove the promoter of this gene in order to silence it, is an applied method and using the precise method of CRISPR/cas9 manifold this efficiency. The remarkable point is that shortcomings of nature should be removed by nature itself. Genetic disorders and some types of cancers are the results of defects in nature, and CRISPR is a system in the nature that can compensate them. However, in the forthcoming study, 2 appropriate gRNAs for the removal of the telomerase gene promoter were presented in silico, but further approvals for this thread require extensive research and in vivo testing.
ACKNOWLEDGEMENT
We appreciate Dr. Mohammad Ali Khosravi and Iran Pasteur Institute because of their genome editing system, CRISPR/Cas9, workshop and working around bioinformatics related soft wares.
REFERENCES
- Lodish (2016) Molecular cell biology. In: (6th edn). Chapter 24.
- Martincorena (2017) Universal Patterns of selection in cancer and somatic tissues. Cell 171(5): 1029-1041.
- Bruce A (2014) Molecular Biology of the Cell. In: (6th edn). Chapter 20.
- Axel AN, Roger R (2002) Telomere maintenance and cancer look, no telomerase. Nature Reviews 2(11): 879-84.
- Kim NW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266(5193): 2011-2015.
- Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33(5): 787–791.
- Hiyama E, Hiyama K (2003) Telomerase as tumor marker. Cancer Lett 194(2): 221-233.
- Calvin B, Harley (2008) Telomerase and cancer therapeutics. Nature Reviews. Volume 8(3): 167-179.
- Phatak P, Burger AM (2007) Telomerase and its potential for therapeutic intervention. Br J Pharmacol 152(7): 1003-1011.
- Armanios M, Greider C W (2005) Telomerase and cancer stem cells. Cold Spring Harb Symp Quant Biol 70: 205-208.
- Phatak P (2007) Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br J Cancer 96: 1223–1233.
- Cortez Gonzalez X, Zanetti M (2007) Telomerase immunity from bench to bedside: round one. J Transl Med 5: 12.
- Thomas G, Charles AG, Carlos F (2013) CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology 31(7): 397-405.
- Mosazadeh M (2016) CRISPR/Cas9 system and cancer. Laboratory diagnosis journal 131: 22-28.
- Bauer DE, Canver MC, Orkin SH (2014) Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J Vis Exp 3(95): e52118.
- Yang H (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154 (6): 1370–1399.
- Wang H (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4): 910–918.
- Shalem O (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343 (6166): 84–87.
- Wang T (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343(6166): 80-84.
- Zhang L, Jia R, Palange NJ, Satheka AC, Togo J, et al. (2015) Large Genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS ONE 10(3): e0120396.
- Shaohua Y, Zhiyao H, Chen (2015) CRISPR/Cas9, Mediated Genome editing of epigenetic factors for cancer therapy. Human Gene Therapy 26(7): 463-471.
- Sánchez RF, Jacks T (2015) Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 15(7): 387–395.
- Jinek M (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816–821.
- Tabebordbar M (2015) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351(6271): 407-411.
- Nivya K (2017) CRISPR-Cas9 mediated gene-silencing of the mutant Huntingtin gene in an In vitro model of huntington’s disease. Int J Mol Sci. 18(4): 754.
- Kellie A (2017) Unexpected mutations after CRISPR–Cas9 editing In vivo. Nature Methods 14(6): 547–548.
- Caleb A (2017) Unexpected mutations after CRISPR-Cas9 editing In vivo” are most likely preexisting sequence variants and not nuclease-induced mutations. bioRxiv.
Article Type
Research Article
Publication history
Received Date: February 02, 2022
Published: March 31, 2022
Address for correspondence
Marziyeh Mousazadeh, Department of Nanobiotechnology, Faculty of Biological Sciences, Tarbiat Modares University, Iran
Copyright
©2022 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Marziyeh M, Zahra R. In Silico Tert-Gene Knocked Out Via Crispr/ Cas9 for Studying the Possibility of Curing Cancerous Cells. 2022- 4(2) OAJBS.ID.000426.
Figure 1: Cancer incidence and mortality rate in United States [3].
Figure 2: Overview of changes in cells that cause cancer [1].
Figure 3: Expression of TERT-gene ratio in different organs.
Figure 4: Different methods by which scientists knocked out the TERT-gene or its product, telomerase [11].
Figure 5: The promoter region of TERT-gene.
Figure 6: The first webpage of CRISPOR online software.
Figure 7: Mentioned PAMs at the 5’ end of TERT-gene promoter.
Figure 8: Mentioned PAMs at the 3’ end of TERT-gene promoter.
Figure 9: Mentioned PAMs at the downstream of TERT-gene promoter.
Table 1: Percentage of somatic mutations in the promoter of telomerase gene.
Table 2: PAMs in 5’ region of TERT-gene promoter (with high specificity).
Table 3: PAMs in 3’ region of TERT-gene promoter (with high specificity).
Table 4: PAMs in 3’ region of TERT gene promoter’s downstream (with high specificity).