Expression, Regulation and Functions of MicroRNAs in Autoimmune Hepatitis
ABSTRACT
Autoimmune hepatitis (AIH) is a rare severe liver disorder, which affects both children and adults worldwide. Studies have shown that AIH may result from the abnormal body’s immune system, which attacks own liver, and causes inflammation and liver damage. However, the exact cause of AIH is unclear. Recent studies have demonstrated that epigenetic regulations including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), DNA methylation and histone modifications play an important role in disease pathogenesis. In this review, expression, regulation and functions of miRNAs in AIH were discussed. It has been shown that specific miRNAs such as miR-21, miR-29a, miR-122, miR-155, miR-221 and miR-223 are involved in the control of cytokine production and regulation of proliferation, apoptosis and functions of liver-infiltrating mononuclear cells, which may be responsible for AIH development. The results may provide insights into the basic mechanisms underlying the dysregulation of miRNAs in AIH that could lead to development of strategies for epigenetic and pharmacologic intervention. Thus, such studies may lead to establishing a rapid diagnostic and prognostic method for early detection and effective treatment of AIH to prevent liver cancer. However, the underlying mechanisms in the expression, regulations and functions of miRNAs in AIH have not been understood and further studies will be necessary.
KEYWORDS
Autoimmune hepatitis; MicroRNAs; Inflammation; Fibrosis; Cirrhosis
INTRODUCTION
Liver cancer has a global incidence of approximately 850,000 cases and represents the fourth leading cause of cancer-related mortality [1]. In 2017, 40,710 new liver cancer cases and 28,920 liver cancer deaths occurred in the United States [2]. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for 80%-85% of all cases of primary liver cancer. Overall HCC incidence rates are increasing in recent years. HCC is the fifth most common cancer, and the third cause of mortality globally [3]. The lack of effective therapeutics in liver cancers and the high failure rate of the vast majority of drugs to date led some investigators to hypothesize that HCC is “not druggable” [4]. HCC occurs most often in people with chronic liver diseases, such as cirrhosis. Cirrhosis is commonly caused by autoimmune hepatitis (AIH), hepatitis B, hepatitis C, alcohol and the other liver disease. Thus, hepatitis and HCC are a tremendous burden for health-care systems and constitute a major challenge to clinicians.
Etiology and Heterogeneity of Hepatitis
Hepatitis refers to inflammation in the liver tissue. Some people with hepatitis may have no symptoms, but others develop yellow discoloration of the skin and whites of the eyes, poor appetite, vomiting, tiredness, abdominal pain and diarrhea. Hepatitis may be acute or chronic. Acute hepatitis lasts for less than six months, and chronic hepatitis exists for more than six months. Acute hepatitis may resolve on its own, progress to chronic hepatitis, or rarely result in acute liver failure. However, chronic hepatitis may progress to scarring of the liver, liver failure, or liver cancer. It has been reported that approximately 10.0% of patients with chronic hepatitis developed cirrhosis, and 20.3% of the cases with compensated cirrhosis progressed to decompensated cirrhosis during a 5-year period. Ultimately, 6.5% of the people with cirrhosis and chronic hepatitis progressed to HCC [5].
The most common cause of hepatitis worldwide is viral infection. There are five main types of viral hepatitis: type A, B, C, D, and E. It has been reported that more than 240 million individuals worldwide are infected with chronic hepatitis B virus (HBV) [5]. Among individuals with chronic HBV infection who are untreated, 15-40% progress to cirrhosis, which may lead to liver failure and liver cancer [6]. Hepatitis B and its related disorders are serious public health issues in undeveloped countries. National and regional prevalence ranges from over 10% in Asia to under 0.5% in the United States and Northern Europe. It is estimated that onethird of 350-400 million individuals worldwide infected with HBV reside in China, with 130 million carriers and 30 million chronically infected [7].
AIH is another important cause of hepatitis. AIH is liver inflammation that occurs when your body’s immune system attacks own liver cells. The exact cause of AIH is unclear, but genetic and environmental factors appear to interact over time in causing the disease. It has been reported that acute exacerbation of AIH poses a significant challenge for diagnosis and can mimic acute viral hepatitis especially in absence of autoantibodies and hypergammaglobulinemia [8]. Furthermore, injection of Concanavalin A (ConA) into mice induces serious immune mediated-liver injury, which is similar to human viral infection and AIH [9]. Thus, the mouse model of AIH induced by ConA could be used in research to understand hepatitis pathogenesis.
Pathophysiological Mechanisms of AIH
AIH is a chronic illness of the liver characterized by hepatocellular inflammation and necrosis, leading to liver injury and dysfunction. AIH affects all ages, both sexes and all ethnic groups worldwide [10]. AIH is most prevalent in European populations. It has been reported that the rates of AIH are approximately from 0.1 to 1.9 per 100,000 annually in Caucasian populations of Europe and North America, whereas AIH is considerably less frequent in Japan [11].
Symptoms of AIH may include fatigue, lethargy, malaise, anorexia, nausea, abdominal pain, itching and arthralgia. AIH may progress to fibrosis, cirrhosis, liver cancer or HCC [12-14]. Studies have shown that autoantibodies, particularly antinuclear antibodies (ANA), smooth muscle antibodies (SMA), liver kidney microsome type 1 antibodies and serum immunoglobulin (Ig)G, increased dramatically in AIH patients [15]. Besides, AIH patients display elevations in liver enzymes such as alanine aminotransferase (ALT) and aspartate aminotransferase (AST), interface hepatitis on histology, and lymphocytic infiltration in the liver. Hepatocyte destruction in AIH is mediated by CD4+ T cells and CD8+ T cells [16]. Particularly, Th17 cells, Th22 cells and pro-inflammatory cytokines such as Interleukin 1 (IL-1) and IL-6 are involved in the destructive liver damage [17,18]. Increased IL-6/IL-12 resulted in the activation of STAT3 and STAT4 in hepatic CD4+CD25+ regulatory T cells (Tregs), thus suppressing Foxp3 gene expression to reduce the suppressive function of Tregs in the liver [19]. However, the pathogenesis of AIH has not been completely understood yet.
Both genetic and environmental factors may be involved in the pathogenesis of AIH [20]. Several reports discovered that gene polymorphisms of TNF-α and CTLA-4 and genes encoding human leucocyte antigens may play a role in AIH development [21,22]. Environmental factors such as diet, sunlight exposure, stress, medication (drugs and herbal agents), hygiene and pathogen infection may influence AIH pathogenesis [20,23].
Studies from AIH mouse model using intravenous injection of ConA into mice have shown that active CD4+ T lymphocytes in the spleen and liver infiltration of active CD4+ T lymphocytes were significantly increased in the ConA-treated group. Moreover, the pro-inflammatory cytokines TNF-α, IFN-γ, and IL-6 were elevated in the plasma as compared with the control group. These results were consistent with those from the clinical AIH patients. Therefore, ConA-induced AIH mouse model is decent for studying the mechanisms of AIH [24]. For instance, ConA-induced AIH mouse model was used to find out that activation of TRPV1 vanilloid receptors could induce myeloid-derived suppressor cells (MDSCs) and thus suppressed AIH [25].
Epigenetics is an exciting and emerging new area of research. Epigenetic mechanisms that are independent of genomic approaches have recently been shown to play a major role in immune activities, and consequently development and progression of diseases [26]. The epigenetic processes consist of changes in the noncoding regions of DNA due to methylation as well as posttranslational modification of histones by methylation, acetylation and deacetylation, and induction of non-coding RNAs including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). In this review, comprehensive data were collected from PubMed, ScienceDirect and the Web of Science databases utilizing the keywords “microRNAs”, “inflammation”, “fibrosis”, “cirrhosis” and “autoimmune hepatitis” in order to understand the expression, regulation and functions of miRNAs in autoimmune hepatitis.
Expression of miRNAs in Autoimmune hepatitis
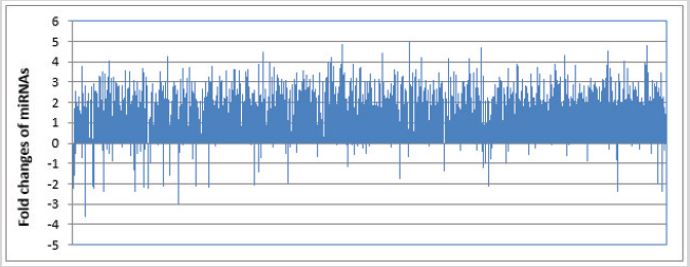
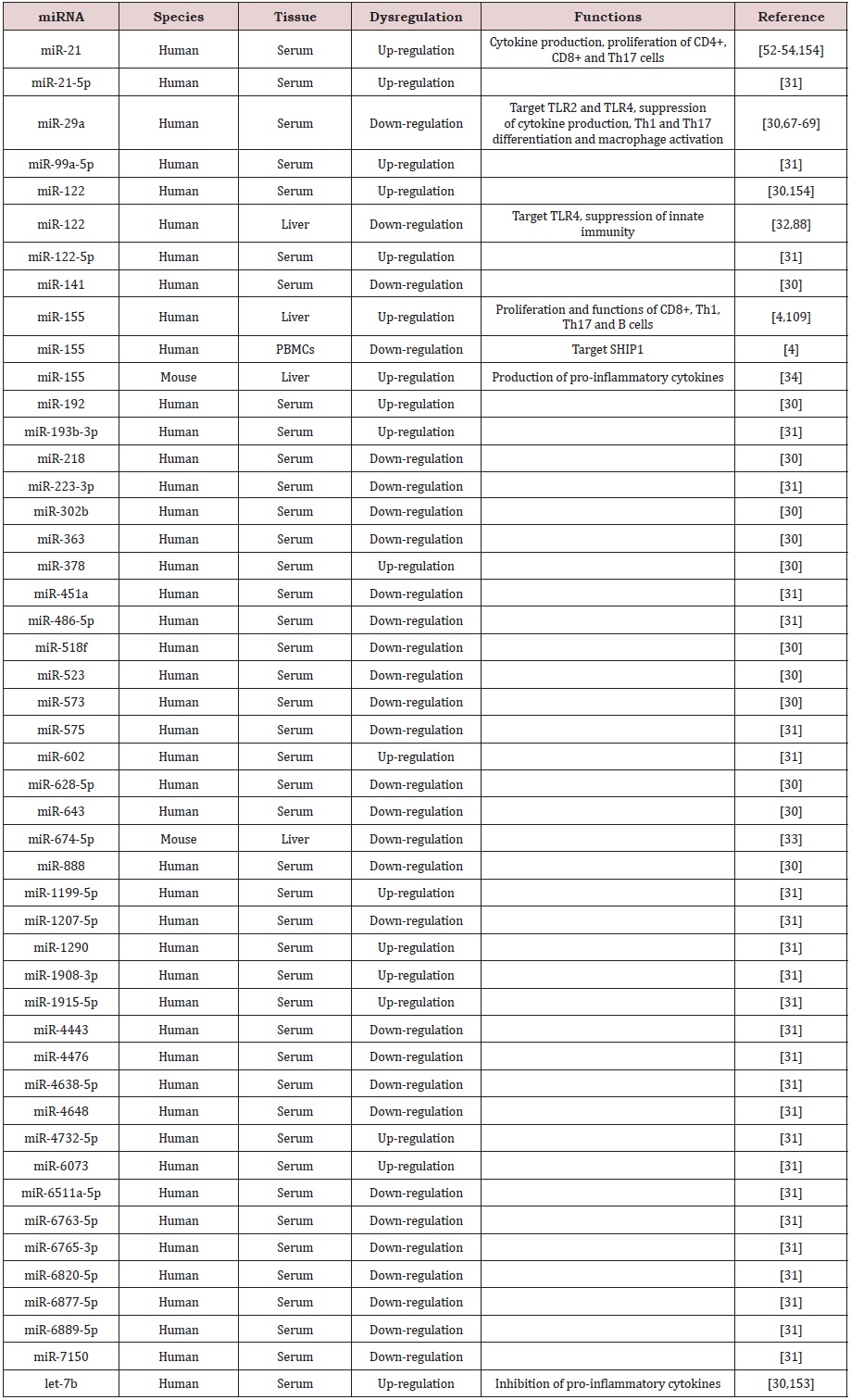
miRNAs are a group of small noncoding RNAs with 19-24 nucleotides in length, which bind to target gene mRNAs via complementary binding to the 3-untranslated region (UTR) and thus regulate gene expression at the post-transcriptional level. Studies have shown that miRNAs are involved in the pathogenesis of liver diseases [27]. It has been reported that there are several liver-specific miRNAs in human, including miR-96, miR-122, miR-130, miR-183, miR-196 and miR-209, which may be used as markers for liver diseases [28,29]. Indeed, quantitative realtime polymerase chain reaction (qRT-PCR) discovered that miR- 122, miR-192, miR-378 and let-7b increased significantly in the circulation of AIH patients, whereas miR-29a, miR-141, miR-218, miR-302b, miR-363, miR-518f, miR-523, miR-573, miR-628-5p, miR-643 and miR-888 significantly decrease in serum samples from AIH patients [30]. Microarray analysis revealed that 11 miRNAs including miR-21-5p, miR-99a-5p, miR-122-5p, miR- 193b-3p, miR-602, miR-1199-5, miR-1290, miR-1908-3p, miR- 1915-5p, miR-4732-5p and miR-6073 were up-regulated, but 16 miRNAs including miR-223-3p, miR-451a, miR-486-5p, miR-575, miR-1207-5p, miR-4443, miR-4476, miR-4638-5p, miR-4648, miR- 6511a-5p, miR-6763-5p, miR-6765-3p, miR-6820-5p, miR-6877- 5p, miR-6889-5p and miR-7150 were down-regulated in the serum samples from patients with type-1 AIH [31]. Interestingly, miR-155 was remarkably up-regulated in the liver, but down-regulated in peripheral blood mononuclear cells (PBMCs) from AIH patients [4]. However, miRNA-122 was significantly down-regulated in the liver biopsy samples, but up-regulated in the serum samples from AIH patients [30,32]. In addition, miR-674-5p was found to be downregulated in Con A-damaged mouse liver [33]. However, miR-155 was up-regulated in liver tissues after 48 h exposure to ConA [34]. Our analysis discovered that many miRNAs including miR-34c-3p, miR-660-3p, miR-1972, 4659a-3p, miR-6832-5p and miR-8068 were up-regulated, but several miRNAs such as miR-122, miR-126, miR-146 and miR-937-3p were down-regulated in AIH mice (Figure 1). Thus, unique miRNAs may be expressed and play an important role in both AIH patients and mouse model, and such miRNAs may be used as biomarkers and therapeutic targets for AIH (Table 1).
Regulation of miRNAs in Autoimmune Hepatitis
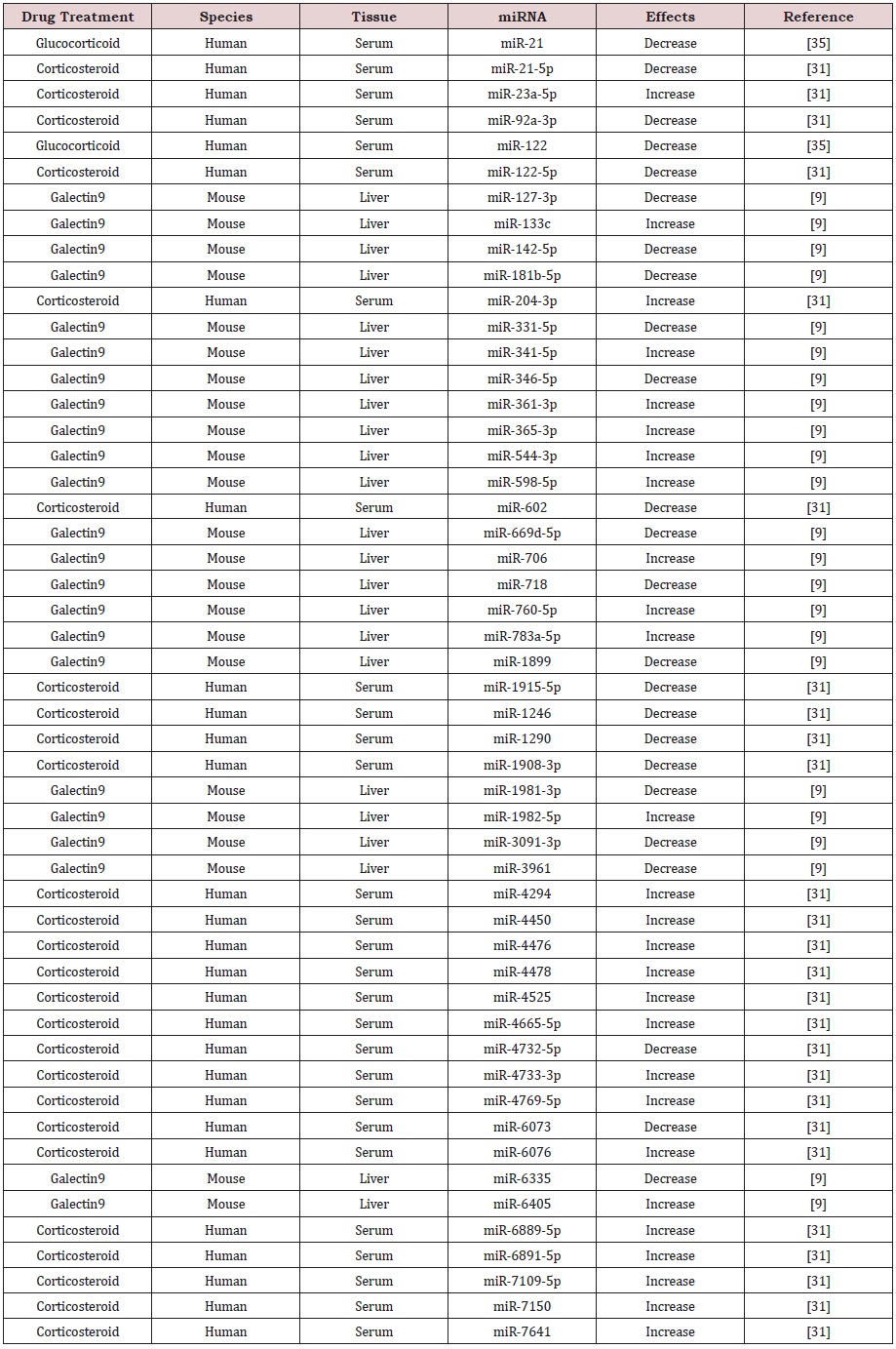
It is interesting to found out that glucocorticoid treatment induced a significant decrease of circulating miR-21 and miR-122 in AIH patients [35]. Other studies discovered that corticosteroid therapy led to not only the up-regulation of 16 miRNAs including miR-23a-5p, miR-204-3p, miR-4294, miR-4450, miR-4476, miR- 4478, miR-4525, miR-4665-5p, miR-4733-3p, miR-4769-5p, miR- 6076, miR-6889-5p, miR-6891-5p, miR-7109-5p, miR-7150 and miR-7641, but also the down-regulation of 10 miRNAs such as miR- 21-5p, miR-92a-3p, miR-122-5p, miR-602, miR-1915-5p, miR-1246, miR-1290, miR-1908-3p, miR-4732-5p and miR-6073 in the sera of patients with type-1 AIH [31]. Following treatment of galectin9, a potential therapeutic agent for AIH, 11 miRNAs including miR-133c, miR-341-5p, miR-361-3p, miR-365-3p, miR-544-3p, miR-598-5p, miR-706, miR-760-5p, miR-783a-5p, miR-1982-5p and miR-6405 were significantly up-regulated, but 12 miRNAs including miR- 127-3p, miR-142-5p, miR-181b-5p, miR-331-5p, miR-346-5p, miR- 669d-5p, miR-718, miR-1899, miR-1981-3p, miR-3091-3p, miR- 3961 and miR-6335 were significantly down-regulated in liver cells from mice challenged with Con A [9]. These results suggest that therapeutic agents may regulate the expression of specific miRNAs and thus play a role in the treatment of AIH, and specific miRNAs may probably be ideal therapeutic targets for AIH (Table 2).
Functions of miRNAs in Autoimmune Hepatitis
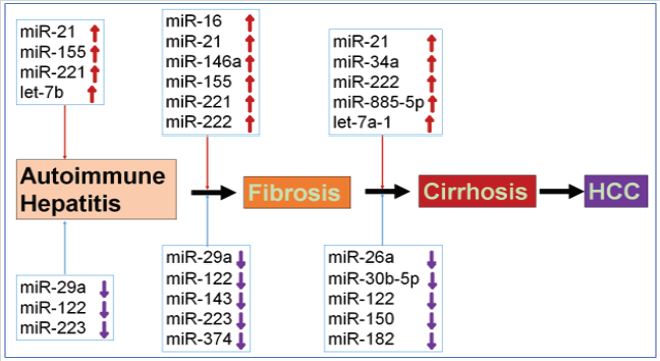
It has been reported that inhibition of miR-155 by antagomir-155 blocked Th17-cells-mediated production of pro-inflammatory cytokines IL-17A and IL-23, and thus reduced ConA-induced liver injury in mice [34]. The results indicated that microRNA-155 may be a fascinating therapeutic target of AIH. In addition, exosomal miR-223 could prevent liver injury in an AIH experimental model [36]. Limited data suggest that specific miRNAs indeed are good targets for AIH treatment. Interestingly, our analysis using the Ingenuity Pathway Analysis® software (IPA; Ingenuity Systems, www.ingenuity.com) discovered that several up-regulated miRNAs such as miR-122b-3p, miR-503-3p and miR-743a-5p in ConAinduced mice could target β-catenin, suggesting that miRNAs may regulate Wnt/β-catenin signaling pathway and thus affect AIH pathogenesis (Figure 2). Moreover, the other up-regulated miRNAs such as miR-124-5p, miR-126a-5p, miR-137-3p and miR- 140-5p could target CASP3, whereas up-regulated miRNAs such as miR-23a-3p, miR-182-5p, miR-204-5p, miR-330-5p and miR- 345-5p could target BCL2, indicating that miRNAs may regulate cell apoptosis and thus affect AIH pathogenesis in mice (Figure 2). These results suggest that specific miRNAs may be expressed and play a role in AIH progression (Figure 3). Below, we discuss the individual roles played by the different miRNAs involved in AIH.
miR-16
miR-16 is a member of miR-15 gene family [37]. miR-16 could inhibit the expression of PD-L1 and thus induced M1 differentiation of mouse peritoneal macrophages, which activate purified CD4+ T cells in vitro [38]. It has also been reported that miR-16 could target and inhibit the expression of the silencing mediator for retinoid and thyroid hormone receptor (SMRT), leading to the activation of NF-κB signaling pathway and increased production of pro-inflammatory cytokines including IL-8, IL-6, and IL-1α [39]. In addition, lipopolysaccharide (LPS) could up-regulate miR-16 and decrease SMAR expression, and thus induced inflammatory liver injury in mice [39,40]. Furthermore, serum levels of miRNA-16 were significantly increased in chronic hepatitis C (CHC) patients with early and late stages of liver fibrosis, suggesting that miR-16 could be used in the detection of liver fibrosis [41]. Therefore, it is possible that miR-16 may play a role in AIH.
miR-21
miR-21 was one of the first mammalian microRNAs identified and target many genes such as BCL2, IL-12p35 and PTEN [42,43]. Serum levels of miR-21 were significantly down-regulated as fibrosis progressed from early fibrosis to late fibrosis in the patients with a chronic HBV infection, suggesting that miR-21 could be used as a biomarker for fibrosis progression [44]. However, Demerdash et al. [45] reported that plasma miR-21 levels increased continuously with the progression of liver fibrosis and development of HCC. miR- 21 could be used as a potential biomarker for fibrosis progression and early detection of HCC [45,46]. miR-21 was significantly increased in surgically removed formalin-fixed paraffin-embedded liver samples from patients with cirrhosis and HCC, and the upregulation of miR-21 in the liver was associated with the progression of cirrhosis and HCC [47]. miR-21 may target smad family member 7 (SMAD7) and thus promote the proliferation and invasion of HCC cells [48]. Therefore, miR-21 may be used as diagnostic marker for HCC [49]. As compared with CHC patients and healthy controls, circulating levels of miR-21 were significantly increased in AIH patients and significantly correlated with the histological grades of inflammation in AIH [31]. It has been reported that miR-21 targeted MyD88 and interleukin-1 receptor-associated kinase 1(IRAK1), and thus was involved in the regulation of immune system and inflammatory response [50,51]. miR-21 could regulate the PTEN/ Akt pathway and activate CD4+ and CD8+ T cells; miR-21 knockout in mice suppressed the proliferation of both CD4+ and CD8+ T cells and deceased their cytokine production [52]. The overexpression of miR-21 increased Th17 cell differentiation, but decreased the production of Treg cells [53,54]. These reports suggest that up upregulated miR-21 plays a central role in AIH pathogenesis by means of the regulation of cytokines, CD4+ T cells, CD8+ T cells, Th17 and Treg cells.
miR-26a
It has been shown that miR-26a is usually down-regulated in cancer cells and tumor tissues [55,56]. miR-26a may target c-Myc, cyclin D2, cyclin E2, cyclin-dependent kinase 4 (CDK4), CDK6, PTEN and thus play an inhibitory roles in cancer cell growth, proliferation, survival, metastasis and invasion [56-58]. miR-26a also plays a role in the regulation of immune responses. It has been reported that miR-26a directly targeted suppressor of cytokine signaling 5 (SOCS5) and ubiquitin-specific protease 3 (USP3), and thus induced the phosphorylation of STAT1 and activated type I IFN signaling pathway and production of IFN-stimulated genes [59,60]. miR-26a could inhibit the function of autoreactive T cells and increased the differentiation of Treg cells in vivo and in vitro [61]. miR-26a also targeted the transcription factor KLF4 and thus regulate the innate immune signaling [62]. Serum miR-26a levels were significantly down-regulated in patients with severe cirrhosis and negatively correlated with severity of cirrhosis in chronic HBV patients [63]. The expression and role of miR-26a in AIH have not been clear so far.
miR-29a
miR-29a was previously known as miR-29. miR-29 family has three members, miR-29a, miR-29b and miR-29c. It has been well documented that miR-29a targets many genes such as CDK2, MUC1and PTEN and is involved in the control of cancer cell proliferation, apoptosis, metastasis and carcinogenesis [64]. miR-29a also play an important role in the regulation of immune responses [65]. For instance, miR-29a could target interferon alpha and beta receptor subunit 1 (IFNAR1) and inhibit IFNAR1 expression, leading to the inhibition of type-I IFN-dependent immune responses [66]. Serum levels of miR-29a were significantly down-regulated as fibrosis progressed from early fibrosis to late fibrosis in the patients with a chronic HBV infection, suggesting that miR-29a could be used as a biomarker for fibrosis progression [44]. It has been reported that miR-29a could inhibit TLR2 and TLR4 signaling pathways and thus protect against liver fibrosis [67]. TLR2 and TLR4 signaling pathways played a critical in cytokine production, Th1 and Th17 differentiation and macrophage activation [68,69]. In the patients with AIH, miR-29a was downregulated in the serum samples [30]. Down-regulation of miR-29a may increase immune responses and promote disease progression of AIH to liver fibrosis.
miR-30b-5p
miR-30b-5p or miR-30b has been shown to be a tumorsuppressor miRNA but acts as an oncogene in some cancers [70,71]. miR-30b-5p could target MTDH and HOXA1 and thus inhibit cancer cell growth, proliferation and invasion [72]. miR-30b-5p targeted SOCS3 and thus suppressed the production of cytokines such as IL- 6, IL-8 and TNF-α, leading to the attenuation of acute lung injury inflammation [73]. miR-30b-5p may also target IL-6R. Compared with hepatitis B patients with liver fibrosis, miR-30b-5p decreased, but IL-6R increased in the patients with liver cirrhosis [74]. Thus, miR-30b-5p may be involved in the progression from liver fibrosis to cirrhosis. In addition, miR-30b-5p could target Nod-like receptor (NLR) family pyrin domain containing 3 (NLRP3) and thus inhibit liver inflammation in the injured liver [75]. Therefore, miR-30b-5p may play a role in AIH pathogenesis.
miR-34a
miR-34a was considered as a tumor suppressor miRNA and played a role in cancer and cardiovascular disease [76,77]. miR- 34a could target BCL-2 and increase apoptosis. miR-34a targeted leucine rich repeat containing G protein-coupled receptor 4 (LGR4), induced GSK-3β-induced p65 serine 468 phosphorylation, activated the NF-κB signaling pathway, leading to the increased production of inflammatory chemokines and cytokines, enhanced inflammation and delayed wound healing [78]. miR-34a also targeted multiple genes involved in T cell immune responses [79]. For example, mR- 34a could targeted FoxP3 and inhibit Treg cell differentiation and function, leading to the excessive immune responses; whereas proinflammatory cytokines IL-6 and TNF-α could bind to the promoter of miR-34a and increase miR-34a expression [80]. It has been reported that miR-34a was significantly up-regulated in the serum from patients with cirrhosis as compared with healthy individuals, suggesting that miR-34a could be used in the detection of liver cirrhosis [81]. The result suggested that miR-34a could be involved in AIH pathogenesis.
miR-122
Animal studies has shown that miR-122 is abundantly expressed in the liver and involved in the regulation of fatty-acid metabolism [82,83]. miR-122 may target several genes including ADAM10, ADAM17, CCNG1 and IGF1R and thus inhibit the expression of these target genes [84]. It has been reported that miR-122 was down-regulated in HCC as compared with normal liver and overexpression of miR-122 decreased tumorigenesis of HCC cell lines [85]. Thus, miRNA-122 is considered as a tumor suppressor gene [86].
It has been reported that miR-122 was significantly upregulated in the serum from patients with cirrhosis as compared with healthy individuals [81]. The result suggested that miR- 122 could be used in the detection of liver cirrhosis. Demerdash et al. [45] also reported that plasma miR-122 levels increased continuously with the progression of liver fibrosis and miR-122 could be used as a potential biomarker for fibrosis progression. However, miR-122 was down-regulated in fibrotic liver biopsies and negatively correlated with fibrosis stage [32,87].
miR-122 targeted TLR4 and thus regulated the innate immunity [88]. miR-122 targeted MERTK, FGFR1 and IGF1R, three receptor tyrosine kinases (RTKs) that directly promoted STAT3 phosphorylation, and thus inhibited the phosphorylation (Tyr705) of STAT3, leading to the suppression of the RTKs/STAT3 signaling pathway and hepatocyte innate immunity [89]. In patients with AIH, miRNA-122 was significantly down-regulated in the liver biopsies, but increased in the serum [30,32]. Serum levels of miR- 122 were also significantly down-regulated in AIH patients with liver cirrhosis and were inversely correlated with increased stages of fibrosis [31]. Therefore, miR-122 could play a protective role in the control of AIH pathogenesis.
miR-143
miR-143 has been found to play an important role in angiogenesis, cardiac morphogenesis and tumorigenesis [90-92]. miR-143 is also involved in inflammation and infection [92]. For instance, miR-143 targeted glucose transporter 1 (Glut-1) and overexpression of miR-143 could significantly promote central memory T CD8+ cell differentiation, suppress cell apoptosis, and increase pro-inflammatory cytokine production [93]. Serum levels of miR-143 were significantly down-regulated as fibrosis progressed from early fibrosis to late fibrosis in the patients with a chronic hepatitis B virus (HBV) infection, suggesting that miR-143 could be used as a biomarker for fibrosis progression [44]. Therefore, miR-143 may play a role in AIH pathogenesis.
miR-146a
It has been shown that miR-146a (previous miR-146) play a critical role in the regulation of inflammation and other process involved in the innate immune system [94]. It has been reported that miR-146a induced Th1-biased immune response [95]. miR- 146a could target tumor-necrosis factor receptor-associated factor 6 (TRAF6), leading to the suppression of NF-κB signaling pathway and inhibition of inflammatory cytokines such as IL-6, IL-8 and TNF-α [96]. miR-146a also targeted STAT1, and up-regulation of miR-146a decreased NK cell-mediated cytotoxicity and the production of IFN-γ and TNF-α, leading to the suppression of NK cell functions in the liver [97]. miR-146a up-regulated significantly in NK cells from chronic HBV-infected patients and HCC patients as compared with healthy controls [97]. It has also been reported that serum levels of miR-146a were significantly increased in CHC patients with early and late stages of liver fibrosis, suggesting that miR-146a could be used in the detection of liver fibrosis [41]. The results suggested that miR-146a could be involved in the regulation of AIH pathogenesis.
miR-150
miR-150 may target several genes such as Akt1, Akt2, c-Myb and forkhead box O1 (FOXO1) [98-100]. miR-150 has been identified to be associated with cancer and infection diseases such as sepsis [101,102] and miR-150 levels in the plasma could be used as a biomarker of HCC and early sepsis [102,103]. miR-150 also played an important role in hematopoiesis, particularly in the promotion of megakaryocyte, B cell, NK cell and T cell differentiation and functions [104-106]. miR-150 has been found out to be involved in autoimmune diseases such as diabetes, multiple sclerosis and rheumatoid arthritis [106]. As compared with non-cirrhotic HCV patients, serum levels of miR-150 significant down-regulated in cirrhotic HCV patients [107]. However, the expression and function of miR-150 in AIH have not been clear yet.
miR-155
It has been well documented that miR-155 plays a critical role in health and numerous diseases such as cancer, cardiovascular and inflammation diseases [108]. It has been reported that miR- 155 increased the proliferation, survival, cytokine production and functions of B cells, CD8+ T cells, Th1 cells, Th17 cells and follicular T helper cells (Tfh), but decreased the differentiation and functions of Th2 cells [109]. HBeAg could regulate PI3K and NFB signal pathways and thus increase miR-155 expression in macrophages. Consequently, miR-155 targeted BCL-6, SHIP-1 and SOCS1 and increased the production of inflammatory cytokines such as TNF-α and IL-6 in HBeAg-stimulated RAW264.7 cells, leading to liver injury [110]. miR-155 also targeted SOCS1 and thus down-regulation of miR-155 increased SOCS1 expression and decreased IFN-γ production in NK cells from chronic hepatitis B patients [111]. In ConA-induced AIH mouse model, miR-155 was significantly up-regulated in liver tissues after 48 h exposure to ConA. Antagomir-155 suppressed the differentiation of Th17 cells in the livers of AIH mice, inhibited production of pro-inflammatory cytokines IL-17A and IL-23, and attenuated ConA-induced liver injury in mice [34]. It has been reported that miR-155 was significantly up-regulated in the liver but decreased in peripheral blood mononuclear cells from patients with AIH. miR-155 was also found to be associated with liver fibrosis progression [4,112]. The results demonstrated that up-regulated miR-155 may induce proinflammatory responses and thus promote AIH pathogenesis.
miR-182
The expression and function of miR-182 (miR-182-5p) in tumor cells are controversial because it can be a tumor suppressor or an oncogene [113]. In glioblastoma, miR-182 targeted Bcl2- like12 (Bcl2L12) and thus induced tumor cell apoptosis. miR-182 targeted c-Met and thus inhibited the division and growth of tumor cells. Also, miR-182 targeted hypoxia-inducible factor 2α (HIF2A) and thus induced tumor cell differentiation [114]. However, miR- 182 could target FOXO3a and thus promote HCC progression [113]. It has been also reported that miR-182 could target FOXO1 and thus increase the clonal expansion of activated Th cells [115]. Recently, miR-182 was found out to be able to target the surfactant protein D (SP-D) and thus increased the production of intestinal TNF-α and IL-6, leading to intestinal injury in Staphylococcus aureus pneumonia-induced sepsis mice [116]. As compared with noncirrhotic HCV patients, serum levels of miR-182 significant downregulated in cirrhotic HCV patients [107]. These results indicated that miR-182 could be involved in AIH pathogenesis.
miR-221
It has been documented that miR-221 may increase cancer cell proliferation, survival, invasion and tumorigenesis [117]. miR-221 played an important role in the promotion of mast cell proliferation, degranulation, cytokine production and adherence, which were associated with asthma, allergy and mastocytosis [118]. miR-221 could also target SOCS1 and thus increase STAT1 phosphorylation and decrease STAT3 phosphorylation, leading to M1 macrophage polarization and inflammatory responses [119]. It has been reported that serum levels of miRNA-221 were significantly increased in CHC patients with early and late stages of liver fibrosis, suggesting that miRNA-221 could be used in the detection of liver fibrosis [41]. miR-221 was also up-regulated in patients with autoimmune diseases [120]. Therefore, it is possible that miR221 may be associated with AIH pathogenesis.
miR-222
miR-222 and miR-221 are two highly homologous miRNAs, which have similar functions in cancer [121]. It has also been reported that serum levels of miRNA-222 were significantly increased in CHC patients with early and late stages of liver fibrosis, suggesting that miRNA-222 could be used in the detection of liver fibrosis [41]. miR-222 was significantly increased in surgically removed formalin-fixed paraffin-embedded liver samples from patients with cirrhosis and HCC, and the up-regulation of miR- 222 in the liver was associated with the progression of cirrhosis and HCC [47]. miR-222 could target the protein phosphatase 2A subunit B (PPP2R2A) and thus activate AKT signaling pathway, leading the progression of HCC [122]. However, the expression and function of miR-222 in immune cells and inflammatory responses have not been understood. miR-222 was up-regulated in patients with autoimmune diseases [120]. miR-222 was also up-regulated in PBMC from sepsis patients and mice, inhibited the expression of a subset of inflammatory genes such as IL-6, IL-12 and TNF and increased immunoparalysis and organ damage [123]. The results suggested miR-222 may play a role in AIH pathogenesis.
miR-223
miR-223 (miR-223-3p) has been shown to play an important role in cancer, infection and autoimmune diseases [124]. miR-223 may also be involved in the regulation of immune responses and autoimmune diseases [125]. It has been reported that miR-223 targeted STAT1, and thus inhibited STAT1 expression and STAT1- dependent signaling pathway in human T cells [126]. miR-223 targeted STAT3 and thus blocked TLR2 and TLR4 signaling pathways and inhibited the production of the pro-inflammatory cytokine IL-6 [127]. miR-223 could regulate neutrophil infiltration, macrophage polarization and inflammasome activation and thus play a role in the pathophysiology of liver diseases [128]. For example, serum levels of miR-223 were significantly down-regulated as liver fibrosis progressed from early fibrosis to late fibrosis in the patients with a chronic HBV infection, suggesting that miR-223 could be used as a biomarker for fibrosis progression [44]. However, Shaker et al. [87] reported that serum levels of miR-223 were significantly up-regulated in HCV patients with significant fibrosis, severe fibrosis and cirrhosis as compared with HCV patients with no or mild fibrosis. Importantly, miR-223 could inhibit the expression of NLRP3 and caspase-1, and thus decrease liver injury in mouse model of AIH [36]. miR-223 could also target STAT3, ameliorate the inflammatory responses and be used in the treatment of AIH [129]. Therefore, miR-223 may play an imperative role in the prevention and treatment of AIH.
miR-374
miR-374 may be involved in many biological processes and disease pathogenesis such as cell growth and differentiation, epilepsy, reproductive disorders, kidney failure and various cancers [130]. miR-374 may also be associated with the regulation of immune responses [131]. Serum levels of miR-374 were significantly down-regulated as fibrosis progressed from early fibrosis to late fibrosis in the patients with a chronic HBV infection, suggesting that miR-374 could be used as a biomarker for fibrosis progression [44]. However, the expression and function of miR-374 in AIH pathogenesis have been unknown so far.
miR-885-5p
It has been reported that miR-885-5p was significantly increased in the sera of patients with liver diseases such as CHB, HCC and liver cirrhosis as compared with normal controls [132]. It is reported that miR-885-5p was significantly down-regulated in HCC tissues and cell lines. miR-885-5p targeted hexokinase 2 (HK2) and thus inhibited glucose uptake, lactate production and glycolysis, leading to the suppression of HCC cell proliferation, growth and migration and induction of apoptosis in vitro and in vivo [133]. miR-885-5p targeted β-catenin CTNNB1 and thus inhibited the activity of Wnt/beta-catenin signaling pathway, leading to the suppression of metastasis of HCC cells in vitro and in vivo and inhibition of HCC progression [134]. miR-885-5p was also down-regulated significantly in neuroblastoma, targeted CDK2 and mini-chromosome maintenance protein (MCM5) and thus induced cell cycle arrest, senescence and apoptosis of neuroblastoma cells [135]. Furthermore, Amaral et al. [81] reported that miR- 885-5p was significantly up-regulated in the serum from patients with cirrhosis as compared with healthy individuals. The result suggested that miR-885-5p could be used in the detection of liver diseases. miR-885-5p also targeted SOCS5, SOCS6 and SOCS7, three suppressor of cytokine signaling genes, suggesting that miR-885- 5p may affect cytokine production [136]. Therefore, miRNA-885-5p may be involved in AIH pathogenesis.
miR-942
miR-942 has been shown to play a promotive role in cancer development [137,138]. For example, miR-942 was significantly upregulated in HCC tissues from HCC patients, targeted ribonucleotide reductase regulatory TP53 inducible subunit M2B (RRM2B) and thus inhibited the expression of the early growth response protein 1 (Egr-1) and PTEN and increased the expression of N-cadherin, MMP-2 and MMP-9, leading to significantly enhancement of HCC cell proliferation, migration, invasion and tumor progression [139]. In addition, TGF-β and LPS induced NF-κB/p50 expression, NF-κB/ p50 bound to the promoter of miR-942 and increased miR-942 expression, miR-942 targeted bone morphogenetic protein and activin membrane-bound inhibitor (BAMBI) and thus promoted liver fibrosis progression [140]. These results suggested that miR- 942 may play a role in AIH pathogenesis. However, the expression and function of miR-942 in immune responses have not been reported so far.
let-7a-1
Lethal-7 (let-7) cluster is composed of let7-a-1, let-7d-1 and let-7f-1. It has been reported that let-7a-1 was significantly upregulated in the serum of HCV liver cirrhosis patients as compared with HCV non-cirrhosis patients [141]. Moreover, let-7a-1 was significantly down-regulated in the serum of HCV-HCC patients as compared with HCV non-HCC cirrhotic patients [141]. The result suggested that let-7a-1 could be used in the detection of liver cirrhosis and HCC. It has also been reported that let-7a-1 could be involved in the promotion of T cell-independent (TI) antigeninduced immunoglobulin (Ig)M antibody production in B cells [142]. Let-7a-1 could target Tet methylcytosine dioxygenase 2 (Tet2) and thus increased IL-6 production in macrophages [143]. These results suggested that let-7a-1 may be associated with AIH pathogenesis.
let-7b
It has been shown that let-7b (let-7b-5p) may play an important role in many biological processes and diseases including immune responses, neural cell differentiation and cancer [144-146]. Let- 7b was found out to be down-regulated in the tumor tissues from different cancer patients [147-149]. Let-7b could target β-catenin and c-Myc and thus inhibit Wnt/β-catenin signaling pathway and suppress HCC cell proliferation [149]. Let-7b could target neuroblastoma RAS viral oncogene homolog (NRAS) and thus inhibit the Ras/Rho signaling pathway, leading to the suppression of prostate cancer cell proliferation, invasion and tumor growth and induction of cancer cell apoptosis [147]. Let-7b could target CEP126 (KIAA1377) and thus inhibit cancer cell proliferation, migration and invasion [150]. It has also been reported that let- 7b could inhibit TGF-β signaling pathway, decrease IFN-γ and IL-4 production, block Th1 and Th2 type cell immune response and inhibit liver fibrosis [151]. Let-7b acted as an endogenous TLR7 ligand and was significantly up-regulated in the liver tissues from alcoholic hepatitis patients and could be responsible for the enhanced inflammatory response in alcoholic hepatitis [152]. Let- 7b could inhibit the production of TNF-α, IL-1β, IL-6 and IL-8 in macrophages [153]. Let-7b could also target TLR4, inhibit the NFĸB signaling pathway and suppress the inflammation and immune responses [154]. Let-7b was found out to be up-regulated in the serum from AIH patients [30]. These results suggested that let-7b could play an important role in AIH pathogenesis.
Prospects of miRNA Regulations in Autoimmune Hepatitis
It has been shown that miRNAs are dysregulated in AIH patients and animal models and drug treatments induce dramatic changes of miRNA expression (Table 1 & 2). Specific miRNAs may also play a role in AIH progression. However, the particular roles and underlying mechanism of dysregulated miRNAs in AIH pathogenesis have not been completely understood. Dysregulated miRNA may be used as diagnostic markers and therapeutic targets for AIH, which require further investigations. In addition, the expression, regulation and functions of other epigenetic mechanisms such as lncRNAs in AIH have not been studies thus far. Furthermore, the changes, functions and underlying mechanisms of DNA methylation and histone modification in AIH have not been known, and further studies will be needed for understanding the epigenetic mechanisms in the regulation and treatment of autoimmune hepatitis.
CONCLUSION
In conclusion, the serum levels of both up-regulated miRNAs such as miR-21, miR-21-5p and miR-122 and down-regulated miRNAs including miR-29a and miR-155 may be used as biomarkers for the diagnosis of autoimmune hepatitis. These miRNAs are also involved in the control of cytokine production and regulation of proliferation, apoptosis and functions of liver-infiltrating mononuclear cells, which may be responsible for AIH development. Thus, these miRNAs may be used as therapeutic targets for autoimmune hepatitis. In addition, the downregulation of miR-21- 5p, miR-92a-3p, miR-122-5p may be used as prognostic markers for the effective treatment of autoimmune hepatitis. The results presented here may provide insights into the basic mechanisms underlying the dysregulation of miRNAs in autoimmune hepatitis that could lead to development of strategies for epigenetic and pharmacologic intervention. Such studies may promisingly lead to establishing a rapid diagnostic and prognostic method for early detection and effective treatment of autoimmune hepatitis to prevent liver cancer.
ACKNOWLEDGEMENT
This work was supported by a grant (#J14LK53 to XC) from the Department of Education of Shandong Province and a grant (#ZR2018MH019 to JZ) from the Natural Science Foundation of Shandong Province, China. The funding sources were not involved in study design, the collection, analysis and interpretation of data, the writing of the report and the decision to submit the article for publication.
REFERENCES
- Li L, Knutsdottir H, Hui K, Weiss MJ, He J, et al. (2019) Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 4: e121490.
- Siegel RL, Miller KD, Jemal A (2017) Cancer Statistics, 2017. CA Cancer J Clin 67: 7-30.
- Taghizadeh S, Alimardani V, Roudbali PL, Ghasemi Y, Kaviani E (2019) Gold Nanoparticles Application in Liver Cancer. Photodiagnosis Photodyn Ther 25: 389-400.
- Blaya D, Aguilar Bravo B, Hao F, Casacuberta Serra S, Coll M, et al. (2018) Expression of microRNA-155 in inflammatory cells modulates liver injury. Hepatology 68: 691-706.
- Qin Y, Zhong Y, Ma T, Zhang J, Yang G, et al. (2017) A pilot study of salivary N-glycome in HBV-induced chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Glycoconj J 34: 523-535.
- Tang LSY, Covert E, Wilson E, Kottilil S (2018) Chronic Hepatitis B Infection: A Review. JAMA 319: 1802-1813.
- Chou FC, Kuo CC, Chen HY, Chen HH, Sytwu HK (2016) DNA demethylation of the TIM-3 promoter is critical for its stable expression on T cells. Genes Immun 17: 179-186.
- Taneja S, Kumar P, Mitra S, Duseja A, Minz R, et al. (2018) Acute exacerbation to autoimmune hepatitis mimicking acute viral hepatitis-A case series and review of literature. J Clin Exp Hepatol 8: 98-103.
- Tadokoro T, Morishita A, Sakamoto T, Fujihara S, Fujita K, et al. (2017) Galectin9 ameliorates fulminant liver injury. Mol Med Rep 16: 36-42.
- Dalekos GN, Koskinas J, Papatheodoridis GV (2019) Hellenic association for the study of the liver clinical practice guidelines: Autoimmune hepatitis. Ann Gastroenterol 32: 1-23.
- Averbukh LD, Wu GY (2018) Role of biologics in the development of autoimmune hepatitis: A review. J Clin Transl Hepatol 6: 402-409.
- Czaja AJ (2014) Review article: The prevention and reversal of hepatic fibrosis in autoimmune hepatitis. Aliment Pharmacol Ther 39: 385-406.
- Krawitt EL (2008) Clinical features and management of autoimmune hepatitis. World J Gastroenterol 14: 3301-3305.
- Tansel A, Katz LH, El Serag HB, Thrift AP, Parepally M, et al. (2017) Incidence and determinants of hepatocellular carcinoma in autoimmune hepatitis: A systematic review and meta-analysis. Clin Gastroenterol Hepatol 15: 1207-1217 e1204.
- Czaja AJ (2011) Performance parameters of the conventional serological markers for autoimmune hepatitis. Dig Dis Sci 56: 545-554.
- Gatselis NK, Zachou K, Koukoulis GK, Dalekos GN (2015) Autoimmune hepatitis, one disease with many faces: etiopathogenetic, clinicolaboratory and histological characteristics. World J Gastroenterol 21: 60-83.
- Ai G, Yan W, Yu H, Xiao F, Xi D, et al. (2018) Soluble Fgl2 restricts autoimmune hepatitis progression via suppressing Tc17 and conventional CD8+ T cell function. J Gene Med 20: e3023.
- Behfarjam F, Jadali Z (2018) Increased expression of aryl hydrocarbon receptor in peripheral blood mononuclear cells of patients with autoimmune hepatitis. Middle East J Dig Dis 10: 105-108.
- Chi G, Feng XX, Ru YX, Xiong T, Gao Y, et al. (2018) TLR2/4 ligand-amplified liver inflammation promotes initiation of autoimmune hepatitis due to sustained IL-6/IL-12/IL-4/IL-25 expression. Mol Immunol 99: 171-181.
- Floreani A, Restrepo Jimenez P, Secchi MF, De Martin S, Leung PSC, et al. (2018) Etiopathogenesis of autoimmune hepatitis. J Autoimmun 95: 133-143.
- Chaouali M, Azaiez MB, Tezeghdenti A, Lagha A, Yacoubi Oueslati B, et al. (2018) Association of TNF-alpha-308 Polymorphism with Susceptibility to Autoimmune Hepatitis in Tunisians. Biochem Genet 56: 650-662.
- Chaouali M, Carvalho A, Tezeghdenti A, Ben Azaiez M, Cunha C, et al. (2018) Cytotoxic T lymphocyte antigen-4 gene polymorphisms and susceptibility to type 1 autoimmune hepatitis in the Tunisian population. Genes Dis 5: 256-262.
- Christen U, Hintermann E (2019) Pathogens and autoimmune hepatitis. Clin Exp Immunol 195: 35-51.
- Ye T, Wang T, Yang X, Fan X, Wen M, et al. (2018) Comparison of concanavalin a-induced murine autoimmune hepatitis models. Cell Physiol Biochem 46: 1241-1251.
- Hegde VL, Nagarkatti PS, Nagarkatti M (2011) Role of myeloid-derived suppressor cells in amelioration of experimental autoimmune hepatitis following activation of TRPV1 receptors by cannabidiol. PLoS One 6: e18281.
- Wilson CB, Rowell E, Sekimata M (2009) Epigenetic control of T-helpercell differentiation. Nat Rev Immunol 9: 91-105.
- Song Y, Yang H, Jiang K, Wang BM, Lin R (2018) miR-181a regulates Th17 cells distribution via up-regulated BCL-2 in primary biliary cholangitis. Int Immunopharmacol 64: 386-393.
- Musaddaq G, Shahzad N, Ashraf MA, Arshad MI (2019) Circulating liverspecific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomarkers 24: 103-109.
- Haider BA, Baras AS, Mc Call MN, Hertel JA, Cornish TC, et al. (2014) A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 9: e89565.
- Yamaura Y, Tatsumi N, Takagi S, Tokumitsu S, Fukami T, et al. (2017) Serum microRNA profiles in patients with chronic hepatitis B, chronic hepatitis C, primary biliary cirrhosis, autoimmune hepatitis, nonalcoholic steatohepatitis, or drug-induced liver injury. Clin Biochem 50: 1034-1039.
- Migita K, Komori A, Kozuru H, Jiuchi Y, Nakamura M, et al. (2015) Circulating microRNA Profiles in Patients with Type-1 Autoimmune Hepatitis. PLoS One 10: e0136908.
- Halasz T, Horvath G, Par G, Werling K, Kiss A, et al. (2015) miR-122 negatively correlates with liver fibrosis as detected by histology and FibroScan. World J Gastroenterol 21: 7814-7823.
- Su K, Wang Q, Qi L, Hua D, Tao J, et al. (2016) MicroRNA-674-5p/5-LO axis involved in autoimmune reaction of Concanavalin A-induced acute mouse liver injury. Toxicol Lett 258: 101-107.
- Xia G, Wu S, Wang X, Fu M (2018) Inhibition of microRNA-155 attenuates concanavalin-A-induced autoimmune hepatitis by regulating Treg/Th17 cell differentiation. Can J Physiol Pharmacol 96: 1293-1300.
- Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C (2017) Role of circulating microRNAs in liver diseases. World J Hepatol 9: 586-594.
- Chen L, Lu FB, Chen DZ, Wu JL, Hu ED, et al. (2018) BMSCs-derived miR- 223-containing exosomes contribute to liver protection in experimental autoimmune hepatitis. Mol Immunol 93: 38-46.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524-15529.
- Jia X, Li X, Shen Y, Miao J, Liu H, et al. (2016) MiR-16 regulates mouse peritoneal macrophage polarization and affects T-cell activation. J Cell Mol Med 20: 1898-1907.
- Zhou R, Li X, Hu G, Gong AY, Drescher KM, et al. (2012) miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One 7: e30772.
- Hamesch K, Borkham Kamphorst E, Strnad P, Weiskirchen R (2015) Lipopolysaccharide-induced inflammatory liver injury in mice. Lab Anim 49: 37-46.
- Abdel Al A, El Ahwany E, Zoheiry M, Hassan M, Ouf A, et al. (2018) miRNA-221 and miRNA-222 are promising biomarkers for progression of liver fibrosis in HCV Egyptian patients. Virus Res 253: 135-139.
- Papagiannakopoulos T, Shapiro A, Kosik KS (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 68: 8164-8172.
- Lu TX, Munitz A, Rothenberg ME (2009) MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 182: 4994-5002.
- Bao S, Zheng J, Li N, Huang C, Chen M, et al. (2017) Serum MicroRNA Levels as a Noninvasive Diagnostic Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Liver Fibrosis. Gut Liver 11: 860-869.
- Demerdash HM, Hussien HM, Hassouna E, Arida EA (2017) Detection of MicroRNA in hepatic cirrhosis and hepatocellular carcinoma in hepatitis C genotype-4 in Egyptian patients. Biomed Res Int 2017: 1806069.
- Dundar HZ, Aksoy F, Aksoy SA, Tasar P, Ugras N, et al. (2019) Overexpression of miR-21 is associated with recurrence in patients with hepatitis B virus-mediated hepatocellular carcinoma undergoing liver transplantation. Transplant Proc 51: 1157-1161.
- Lendvai G, Szekerczes T, Gyongyosi B, Schlachter K, Kontsek E, et al. (2019) MicroRNA expression in focal nodular hyperplasia in comparison with cirrhosis and hepatocellular carcinoma. Pathol Oncol Res 25: 1103- 1109.
- Wang Y, Zhang P, Yuan M, Li X (2019) Overexpression of miRNA-21 promotes the proliferation and invasion in hepatocellular carcinoma cells via suppressing SMAD7. Technol Cancer Res Treat 18: 1533033819878686.
- Qu J, Yang J, Chen M, Cui L, Wang T, et al. (2019) MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak J Med Sci 35: 1466-1471.
- Feng N, Zhou Z, Li Y, Zhao L, Xue Z, et al. (2017) Enterovirus 71-induced has-miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. Virus Res 237: 27-36.
- Sheedy FJ (2015) Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immunol 6: 19.
- He W, Wang C, Mu R, Liang P, Huang Z, et al. (2017) MiR-21 is required for anti-tumor immune response in mice: an implication for its bidirectional roles. Oncogene 36: 4212-4223.
- Jin S, Chen H, Li Y, Zhong H, Sun W, et al. (2018) Maresin 1 improves the Treg/Th17 imbalance in rheumatoid arthritis through miR-21. Ann Rheum Dis 77: 1644-1652.
- Zheng X, Dong L, Wang K, Zou H, Zhao S, et al. (2019) MiR-21 Participates in the PD-1/PD-L1 Pathway-Mediated Imbalance of Th17/Treg Cells in Patients After Gastric Cancer Resection. Ann Surg Oncol 26: 884-893.
- Reuland SN, Smith SM, Bemis LT, Goldstein NB, Almeida AR, et al. (2013) MicroRNA-26a is strongly downregulated in melanoma and induces cell death through repression of silencer of death domains (SODD). J Invest Dermatol 133: 1286-1293.
- Gao J, Liu QG (2011) The role of miR-26 in tumors and normal tissues (Review). Oncol Lett 2: 1019-1023.
- Lu J, He ML, Wang L, Chen Y, Liu X, et al. (2011) MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 71: 225-233.
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, et al. (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005-1017.
- Zhang J, Li Z, Huang J, Yin H, Tian J, et al. (2019) miR-26a Inhibits Feline Herpesvirus 1 Replication by Targeting SOCS5 and Promoting Type I Interferon Signaling. Viruses 12: 2.
- Gao S, Li J, Song L, Wu J, Huang W (2017) Influenza A virus-induced downregulation of miR-26a contributes to reduced IFNalpha/beta production. Virol Sin 32: 261-270.
- Ma H, Zhang S, Shi D, Mao Y, Cui J (2016) MicroRNA-26a Promotes Regulatory T cells and Suppresses Autoimmune Diabetes in Mice. Inflammation 39: 1-9.
- Sahu SK, Kumar M, Chakraborty S, Banerjee SK, Kumar R, et al. (2017) MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPbeta regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog 13: e1006410.
- Jiang S, Jiang W, Xu Y, Wang X, Mu Y, et al. (2019) Serum miR-21 and miR- 26a Levels Negatively Correlate with Severity of Cirrhosis in Patients with Chronic Hepatitis B. Microrna 8: 86-92.
- Wang JY, Zhang Q, Wang DD, Yan W, Sha HH, et al. (2018) MiR-29a: a potential therapeutic target and promising biomarker in tumors. Biosci Rep 38: BSR20171265.
- Tian R, Zheng Z, Huang R, Jiao Y, Du X, et al. (2015) miR-29a Participated in nacre formation and immune response by targeting Y2R in pinctada martensii. Int J Mol Sci 16: 29436-29445.
- Zhang Y, Yang L, Wang H, Zhang G, Sun X, et al. (2016) Respiratory syncytial virus non-structural protein 1 facilitates virus replication through miR-29a-mediated inhibition of interferon-alpha receptor. Biochem Biophys Res Commun 478: 1436-1441.
- Lin YC, Wang FS, Yang YL, Chuang YT, Huang YH, et al. (2018) MicroRNA- 29a mitigation of toll-like receptor 2 and 4 signaling and alleviation of obstructive jaundice-induced fibrosis in mice. Biochem Biophys Res Commun 496: 880-886.
- Li JY, Liu Y, Gao XX, Gao X, Cai H, et al. (2014) TLR2 and TLR4 signaling pathways are required for recombinant Brucella abortus BCSP31- induced cytokine production, functional upregulation of mouse macrophages, and the Th1 immune response in vivo and in vitro. Cell Mol Immunol 11: 477-494.
- Sugitharini V, Shahana P, Prema A, Berla Thangam E (2016) TLR2 and TLR4 co-activation utilizes distinct signaling pathways for the production of Th1/Th2/Th17 cytokines in neonatal immune cells. Cytokine 85: 191-200.
- Le Guillou S, Sdassi N, Laubier J, Passet B, Vilotte M, et al. (2012) Overexpression of miR-30b in the developing mouse mammary gland causes a lactation defect and delays involution. PLoS One 7: e45727.
- Wu T, Song H, Xie D, Hua K, Hu J, et al. (2020) Mir-30b-5p Promotes Proliferation, Migration, and Invasion of Breast Cancer Cells via Targeting ASPP2. Biomed Res Int 2020: 7907269.
- Xu J, Lv H, Zhang B, Xu F, Zhu H, et al. (2019) miR-30b-5p acts as a tumor suppressor microRNA in esophageal squamous cell carcinoma. J Thorac Dis 11: 3015-3029.
- Zhou T, Chen YL (2019) The functional mechanisms of miR-30b-5p in acute lung injury in children. Med Sci Monit 25: 40-51.
- Chen Y, Yang S, Peng Y, Yang Z (2017) The regulatory role of IL-6R in hepatitis B-associated fibrosis and cirrhosis. Braz J Med Biol Res 50: e6246.
- Yang L, Tian L, Zhang Z, Zhou X, Ji X, et al. (2020) Cannabinoid receptor 1/miR-30b-5p axis governs macrophage NLRP3 expression and inflammasome activation in liver inflammatory disease. Mol Ther Nucleic Acids 20: 725-738.
- Zhang L, Liao Y, Tang L (2019) MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res 38: 53.
- Li N, Wang K, Li PF (2015) MicroRNA-34 Family and its role in cardiovascular disease. Crit Rev Eukaryot Gene Expr 25: 293-297.
- Wu J, Li X, Li D, Ren X, Li Y, et al. (2020) MicroRNA-34 family enhances wound inflammation by targeting LGR4. J Invest Dermatol 140: 465-476 e411.
- Hart M, Walch-Ruckheim B, Krammes L, Kehl T, Rheinheimer S, et al. (2019) miR-34a as hub of T cell regulation networks. J Immunother Cancer 7: 187.
- Xie M, Wang J, Gong W, Xu H, Pan X, et al. (2019) NF-kappaB-driven miR- 34a impairs Treg/Th17 balance via targeting Foxp3. J Autoimmun 102: 96-113.
- Amaral AED, Rode MP, Cisilotto J, Silva TED, Fischer J, et al. (2018) MicroRNA profiles in serum samples from patients with stable cirrhosis and miRNA-21 as a predictor of transplant-free survival. Pharmacol Res 134: 179-192.
- Laterza OF, Scott MG, Garrett-Engele PW, Korenblat KM, Lockwood CM, et al. (2013) Circulating miR-122 as a potential biomarker of liver disease. Biomark Med 7: 205-210.
- Chung HH (2018) New insights for controversial issues of miR-122 in hepatic lipid metabolism. Gastroenterology 154: 1552-1553.
- Chen CL, Wu JC, Chen GY, Yuan PH, Tseng YW, et al. (2015) Baculovirusmediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol Ther 23: 79-88.
- Nakao K, Miyaaki H, Ichikawa T (2014) Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol 49: 589-593.
- Wen J, Friedman JR (2012) miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest 122: 2773-2776.
- Shaker OG, Senousy MA (2017) Serum microRNAs as predictors for liver fibrosis staging in hepatitis C virus-associated chronic liver disease patients. J Viral Hepat 24: 636-644.
- Shi L, Zheng X, Fan Y, Yang X, Li A, et al. (2019) The contribution of miR-122 to the innate immunity by regulating toll-like receptor 4 in hepatoma cells. BMC Gastroenterol 19: 130.
- Xu H, Xu SJ, Xie SJ, Zhang Y, Yang JH, et al. (2019) MicroRNA-122 supports robust innate immunity in hepatocytes by targeting the RTKs/STAT3 signaling pathway. Elife 8: e41159.
- Wang R, Zhang H, Ding W, Fan Z, Ji B, et al. (2020) miR-143 promotes angiogenesis and osteoblast differentiation by targeting HDAC7. Cell Death Dis 11: 179.
- Vacante F, Denby L, Sluimer JC, Baker AH (2019) The function of miR- 143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vascul Pharmacol 112: 24-30.
- Roderburg C, Koch A, Benz F, Vucur M, Spehlmann M, et al. (2019) Serum levels of miR-143 predict survival in critically ill patients. Dis Markers 2019: 4850472.
- Zhang T, Zhang Z, Li F, Ping Y, Qin G, et al. (2018) miR-143 regulates memory T cell differentiation by reprogramming T cell metabolism. J Immunol 201: 2165-2175.
- Lee HM, Kim TS, Jo EK (2016) MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep 49: 311-318.
- Hou T, Liao J, Zhang C, Sun C, Li X, et al. (2018) Elevated expression of miR-146, miR-139 and miR-340 involved in regulating Th1/Th2 balance with acute exposure of fine particulate matter in mice. Int Immunopharmacol 54: 68-77.
- Wang XP, Luoreng ZM, Zan LS, Li F, Li N (2017) Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene. J Dairy Sci 100: 7648-7658.
- Xu D, Han Q, Hou Z, Zhang C, Zhang J (2017) miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell Mol Immunol 14: 712-720.
- Chen W, Han C, Zhang J, Song K, Wang Y, et al. (2015) miR-150 deficiency protects against FAS-induced acute liver injury in mice through regulation of AKT. PLoS One 10: e0132734.
- Chen Z, Stelekati E, Kurachi M, Yu S, Cai Z, et al. (2017) miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep 20: 2584-2597.
- Ban YH, Oh SC, Seo SH, Kim SM, Choi IP, et al. (2017) miR-150-Mediated foxo1 regulation programs CD8(+) T cell differentiation. Cell Rep 20: 2598-2611.
- He Z, Dang J, Song A, Cui X, Ma Z, et al. (2020) The involvement of miR-150/beta-catenin axis in colorectal cancer progression. Biomed Pharmacother 121: 109495.
- Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Scholten D, et al. (2013) Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 8(1): e54612.
- Yu F, Lu Z, Chen B, Dong P, Zheng J (2015) MicroRNA-150: a promising novel biomarker for hepatitis B virus-related hepatocellular carcinoma. Diagn Pathol 10: 129.
- He Y, Jiang X, Chen J (2014) The role of miR-150 in normal and malignant hematopoiesis. Oncogene 33(30): 3887-3893.
- Smith NL, Wissink EM, Grimson A, Rudd BD (2015) miR-150 regulates differentiation and cytolytic effector function in CD8+ T cells. Sci Rep 5: 16399.
- Huang XL, Zhang L, Li JP, Wang YJ, Duan Y, et al. (2015) microRNA-150: A potential regulator in pathogens infection and autoimmune diseases. Autoimmunity 48: 503-510.
- Shaheen NMH, Zayed N, Riad NM, Tamim HH, Shahin RMH, et al. (2018) Role of circulating miR-182 and miR-150 as biomarkers for cirrhosis and hepatocellular carcinoma post HCV infection in Egyptian patients. Virus Res 255: 77-84.
- Faraoni I, Antonetti FR, Cardone J, Bonmassar E (2009) miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792(6): 497-505.
- Alivernini S, Gremese E, McSharry C, Tolusso B, Ferraccioli G, et al. (2017) microRNA-155-at the critical interface of innate and adaptive immunity in arthritis. Front Immunol 8: 1932.
- Wang W, Bian H, Li F, Li X, Zhang D, et al. (2018) HBeAg induces the expression of macrophage miR-155 to accelerate liver injury via promoting production of inflammatory cytokines. Cell Mol Life Sci 75: 2627-2641.
- Ge J, Huang Z, Liu H, Chen J, Xie Z, et al. (2017) Lower expression of microRNA-155 contributes to dysfunction of natural killer cells in patients with chronic hepatitis B. Front Immunol 8: 1173.
- Szekerczes T, Gogl A, Illyes I, Mandl J, Borka K, et al. (2020) Autophagy, mitophagy and microRNA expression in chronic hepatitis C and autoimmune hepatitis. Pathol Oncol Res 26: 2143-2151.
- Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY, et al. (2018) miR-182- 5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol 11(1): 12.
- Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, et al. (2015) miR- 182 integrates apoptosis, growth and differentiation programs in glioblastoma. Genes Dev 29(7): 732-745.
- Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, et al. (2010) The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol 11: 1057- 1062.
- Du X, Wei J, Tian D, Wu M, Yan C, et al. (2020) miR-182-5p contributes to intestinal injury in a murine model of Staphylococcus aureus pneumonia-induced sepsis via targeting surfactant protein D. J Cell Physiol 235(1): 563-572.
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, et al. (2010) miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 107(1): 264-269.
- Mayoral RJ, Deho L, Rusca N, Bartonicek N, Saini HK, et al. (2011) MiR- 221 influences effector functions and actin cytoskeleton in mast cells. PLoS One 6(10): e26133.
- Cai M, Shi Y, Zheng T, Hu S, Du K, et al. (2020) Mammary epithelial cell derived exosomal MiR-221 mediates M1 macrophage polarization via SOCS1/STATs to promote inflammatory response. Int Immunopharmacol 83: 106493.
- Abo ElAtta AS, Ali YBM, Bassyouni IH, Talaat RM (2019) Upregulation of miR-221/222 expression in rheumatoid arthritis (RA) patients: correlation with disease activity. Clin Exp Med 19(1): 47-53.
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12(1): 27-33.
- Wong QW, Ching AK, Chan AW, Choy KW, To KF, et al. (2010) MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res 16(3): 867-875.
- Seeley JJ, Baker RG, Mohamed G, Bruns T, Hayden MS, et al. (2018) Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 559: 114-119.
- Haneklaus M, Gerlic M, O’Neill LA, Masters SL (2013) miR-223: infection, inflammation and cancer. J Intern Med 274(3): 215-226.
- Zhang L, Wu H, Zhao M, Lu Q (2020) Identifying the differentially expressed microRNAs in autoimmunity: A systemic review and metaanalysis. Autoimmunity 53(3): 122-136.
- Moles R, Bellon M, Nicot C (2015) STAT1: A novel target of miR-150 and miR-223 is involved in the proliferation of HTLV-I-transformed and ATL cells. Neoplasia 17(5): 449-462.
- Wu J, Niu P, Zhao Y, Cheng Y, Chen W, et al. (2019) Impact of miR-223- 3p and miR-2909 on inflammatory factors IL-6, IL-1ss, and TNF-alpha, and the TLR4/TLR2/NF-kappaB/STAT3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PLoS One 14: e0212063.
- Ye D, Zhang T, Lou G, Liu Y (2018) Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med 50(9): 128.
- Lu FB, Chen DZ, Chen L, Hu ED, Wu JL, et al. (2019) Attenuation of experimental autoimmune hepatitis in mice with bone mesenchymal stem cell-derived exosomes carrying microRNA-223-3p. Mol Cells 42: 906-918.
- Bian H, Zhou Y, Zhou D, Zhang Y, Shang D, et al. (2019) The latest progress on miR-374 and its functional implications in physiological and pathological processes. J Cell Mol Med 23(5): 3063-3076.
- Martins MR, Almeida RS, Lucena-Silva N, Coutinho-Camilo CM, Torjal I, et al. (2019) MicroRNA expression profiling provides novel insights into immune-related pathways involved in gastric cancer. Med Oncol 36(9): 81.
- Gui J, Tian Y, Wen X, Zhang W, Zhang P, et al. (2011) Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 120: 183-193.
- Xu F, Yan JJ, Gan Y, Chang Y, Wang HL, et al. (2019) miR-885-5p negatively regulates warburg Effect by silencing hexokinase 2 in liver cancer. Mol Ther Nucleic Acids 18: 308-319.
- Zhang Z, Yin J, Yang J, Shen W, Zhang C, et al. (2016) miR-885-5p suppresses hepatocellular carcinoma metastasis and inhibits Wnt/ beta-catenin signaling pathway. Oncotarget 7(46): 75038-75051.
- Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, et al. (2011) microRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ 18: 974- 984.
- Su M, Qin B, Liu F, Chen Y, Zhang R (2018) miR-885-5p upregulation promotes colorectal cancer cell proliferation and migration by targeting suppressor of cytokine signaling. Oncol Lett 16(1): 65-72.
- Yang F, Shao C, Wei K, Jing X, Qin Z, et al. (2019) miR-942 promotes tumor migration, invasion, and angiogenesis by regulating EMT via BARX2 in non-small-cell lung cancer. J Cell Physiol 234(12): 23596- 23607.
- Zhang J, Zhang Z, Sun J, Ma Q, Zhao W, et al. (2019) MiR-942 regulates the function of breast cancer cell by targeting FOXA2. Biosci Rep 39: BSR20192298.
- Zhang Q, Zhu B, Qian J, Wang K, Zhou J (2019) miR-942 promotes proliferation and metastasis of hepatocellular carcinoma cells by inhibiting RRM2B. Onco Targets Ther 12: 8367-8378.
- Tao L, Xue D, Shen D, Ma W, Zhang J, et al. (2018) microRNA-942 mediates hepatic stellate cell activation by regulating BAMBI expression in human liver fibrosis. Arch Toxicol 92: 2935-2946.
- Aly DM, Gohar NA, Abd El-Hady AA, Khairy M, Abdullatif MM (2020) Serum microRNA let-7a-1/let-7d/let-7f and miRNA 143/145 Gene Expression Profiles as Potential Biomarkers in HCV Induced Hepatocellular Carcinoma. Asian Pac J Cancer Prev 21: 555-562.
- Jiang S, Yan W, Wang SE, Baltimore D (2018) Let-7 suppresses B cell activation through restricting the availability of necessary nutrients. Cell Metab 27(2): 393-403.
- Jiang S, Yan W, Wang SE, Baltimore D (2019) Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc Natl Acad Sci U S A 116(25): 12416-12421.
- Teng GG, Wang WH, Dai Y, Wang SJ, Chu YX, et al. (2013) Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting toll-like receptor 4. PLoS One 8: e56709.
- Zhao C, Sun G, Li S, Lang MF, Yang S, et al. (2010) MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A 107(5): 1876- 1881.
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M (2008) MicroRNA let- 7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18(5): 549-557.
- Huang L, Wang L, Wu Z, Jiang F, Li Q, et al. (2020) MicroRNA let-7b Inhibits Proliferation and Induces Apoptosis of Castration-Resistant Prostate Cancer Cells by Blocking the Ras/Rho Signaling Pathway via NRAS. Clin Transl Sci.
- Bozgeyik E (2020) Bioinformatic analysis and in vitro validation of Let-7b and Let-7c in breast cancer. Comput Biol Chem 84: 107191.
- Wang Y, Mo Y, Wang L, Su P, Xie Y (2018) Let-7b contributes to hepatocellular cancer progression through Wnt/beta-catenin signaling. Saudi J Biol Sci 25(5): 953-958.
- Zheng S, Liu Q, Ma R, Tan D, Shen T, et al. (2019) Let-7b-5p inhibits proliferation and motility in squamous cell carcinoma cells through negative modulation of KIAA1377. Cell Biol Int 43(6): 634-641.
- Tang N, Wu Y, Cao W, Liang Y, Gao Y, et al. (2017) Lentivirus-mediated over-expression of let-7b microRNA suppresses hepatic fibrosis in the mouse infected with Schistosoma japonicum. Exp Parasitol 182: 45-53.
- Massey VL, Qin L, Cabezas J, Caballeria J, Sancho-Bru P, et al. (2018) TLR7-let-7 Signaling Contributes to Ethanol-Induced Hepatic Inflammatory Response in Mice and in Alcoholic Hepatitis. Alcohol Clin Exp Res 42(11): 2107-2122.
- Nematian SE, Mamillapalli R, Kadakia TS, Majidi Zolbin M, Moustafa S, et al. (2018) Systemic Inflammation Induced by microRNAs: Endometriosis-Derived Alterations in Circulating microRNA 125b- 5p and Let-7b-5p Regulate Macrophage Cytokine Production. J Clin Endocrinol Metab 103(1): 64-74.
- Czaja AJ (2018) Epigenetic changes and their implications in autoimmune hepatitis. Eur J Clin Invest 48: e12899.
Article Type
Review Article
Publication history
Received date: October 22, 2020
Published date: November 10, 2020
Address for correspondence
Xiaoyan Chi, MSc, Institute for Tumor Immunology,
Ludong University School of Life Sciences, 186 Hongqi Middle Road, Yantai, PR
China
Juhua Zhou, PhD, Institute for Tumor Immunology, Ludong University School of Life
Sciences, Yantai, PR China
Copyright
©2020 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Xiaoyan C, Qingrong H, Yanmin L, Xiaoguang Y, Jiang F, Juhua Z. Expression, Regulation and Functions of MicroRNAs in Autoimmune Hepatitis. 2020 - 2(6) OAJBS.ID.000234.
Figure 1: Change pattern of miRNA expression in Con A-induced autoimmune hepatitis mice. Liver tissues were collected from C57BL/6 mice at 16 h after Con A injection. Liver tissues were used in the preparation of total RNA samples and then the total mRNA samples were used in RNA sequencing by Illumina NextSeq 500 system. The sequence data were analyzed using the Galaxy web-based platform (https://usegalaxy.org/). Based on the changes of gene expression, the changes of expression level of miRNAs were determined using the Ingenuity Pathway Analysis® software (IPA; Ingenuity Systems, www.ingenuity.com). Each vertical line represented a single miRNA in the figure.
Figure 2: Pathway analysis of miRNAs dysregulated in Con A-induced autoimmune hepatitis mice. The expression changes of miRNAs dysregulated in liver tissues from Con A-injected mice were determined by RNA sequencing. The pathways of targets of miRNAs dysregulated in Con A-injected mice were analysed using the Ingenuity Pathway Analysis® Platform (IPA; Ingenuity Systems, www.ingenuity.com). The pathways of β-catenin (A), CASP3 (B) and BCL2 (C) targeted by miRNAs dysregulated in Con A-injected mice were presented. miRNAs marked in red represented the up regulation, whereas those marked in green represented the downregulation.
Figure 3: Expression and function of miRNAs in autoimmune hepatitis progression. Autoimmune hepatitis may lead to the development of fibrosis, and further progress to cirrhosis and HCC. Specific miRNAs were dysregulated and played an important role in the progress of autoimmune hepatitis. Upwards arrows represented the up regulation of specific miRNAs, and downwards arrows represented the downregulation of specific miRNAs in the stage of autoimmune disease progress.
Table 1: Dysregulation of miRNAs in autoimmune hepatitis.
Table 2: Effects of drug treatments on the expression of miRNAs in autoimmune hepatitis.