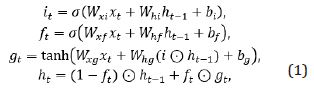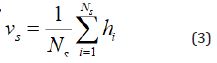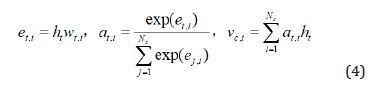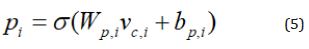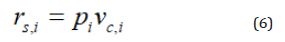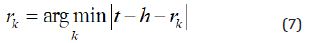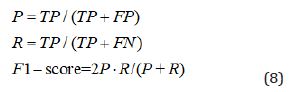Drug-Drug Interaction Extraction Based on Bidirectional Gated Recurrent Unit networks and Capsule Networks
ABSTRACT
Drug-drug interactions (DDIs) often result in unexpected side effects even death. DDI extraction (DDIE) is an important topic in the field of biomedical relationship extraction and has attracted great attention in natural language processing. The existing methods mainly emphasize the influence of entity, location and other information on DDIE, the maximum pooling of convolutional neural network (CNN) and Recurrent Neural Network results in the loss of feature information, and their extraction performance in long and complex sentences is still unsatisfactory. In this paper, a Gated Recurrent Unit and Capsule Networks (GRUTC) model is constructed for DDIE. Firstly, the original sentence sequence and the shortest dependency path (SDP) sequence are computed from the corpus and are input into two independent bidirectional Gated Recurrent Unit networks (Bi-GRU) to learn its semantic information and context information. Secondly, the extracted feature vectors are concatenated and input to the capsule network. Aiming at the problem of information loss caused by pooling strategy of CNN, the dynamic routing mechanism is employed to dynamically determine the amount of information transferred from low-level capsules to high-level capsules, so as to make full use of the high-level characteristic information and improve the DDIE effect. The experimental results on DDIExtraction2013 corpus show that the proposed method can effectively improve the performance without using any artificial features, and the F1-score is 73.7%, reaching the current advanced level.
KEYWORDS
DDI: Drug-drug Interactions; DI: Dependency Information; Bi-GRU: Bidirectional Gated Recurrent Unit networks; CN: Capsule Networks
INTRODUCTION
Drug-drug interactions (DDI) are the pharmacological interactions of one drug to another when one patient takes more than one kind of drug simultaneously or at a certain time. DDIs may be strengthened or weakened the treatment effect or may lead to various adverse DDIs (ADDIs), which will bring inevitable harmful consequences to the patient. ADDIs are serious health hazards and sometimes even result in death [1]. It is reported that more than 300,000 deaths are caused by ADDIs per year in the USA and Europe [2]. According to data from Centers for Disease Control and Prevention, the number of ADDIs harm are anywhere from 1.9 to 5 million inpatients per year. Owing to the aging of population taking multiple medications, the ADDI problem likely continues to get worse. As a result, DDIE has been taken seriously by pharmaceutical companies and drug agencies in drug safety and healthcare management. There are some DDI databases such as Drug Bank, PharmGKB and KEGG and a large number of DDI findings reported in unstructured biomedical literature [3,4]. These DDIs can help physicians and pharmacists avoid the common ADDIs, but it is known that clinical studies cannot sufficiently andaccurately identify DDIs for new drugs before they are available on the market, and the existing public and proprietary sources of DDI information are known to be incomplete and/or inaccurate and so not reliable. Therefore, extracting DDI during drug development can reduce ADDI and development costs and time by rigorously evaluating drug candidates. DDI is a kind of inter-entity relationship extraction task with multi-classification and no distinction of relationship direction.
The study of DDI can provide more in-depth information for the construction and maintenance of biomedical database, and provide more important reference for disease treatment, drug development and life science research [5,6]. Vilar et al. [7] proposed a DDIs prediction method based on the similarity of DDI candidates to drugs involved in established DDIs. The method integrates a reference standard database of known DDIs with drug similarity information extracted from different sources, such as 2D and 3D molecular structures, interaction profiles, targets and side-effect similarities. Mezaache et al. [8] assessed the incidence of ADDIs related to immune thrombocytopenia drugs and compared the incidence of ADDIs depending on the drugs, and evaluated the factors associated to corticosteroids related ADDI occurrence. Kim et al. [9] extracted DDIs from literature using a rich feature-based linear kernel approach, including word features, word pair features, analytic tree features, and noun phrase consistency. To predict side effects of new drugs, Zhang et al. [10] defined a drug-drug similarity for DDIE by exploring linear neighbourhood relationship. They transferred the similarity from the feature space into the side effect space and predicted drug side effects by propagating known side effect information through a similarity-based graph. In the current study, all public available sources of potential DDI information that could be identified using a comprehensive and broad search were combined into a single dataset. The most of traditional DDIE approaches mainly rely on support vector machines (SVM) with a large number of manually defined features, but it is difficult to extract the optimal classification features due to the complexity of NLP.
Recently, deep learning has achieved breakthroughs in modeling complex structures in different NLP tasks, and has been widely used in DDIE. Liu et al. [11] proposed a CNN-based method for DDIE. The experiments conducted on the DDIExtraction2013 challenge corpus demonstrated that CNN is a good choice for DDI extraction. Zhao et al. [12] presented a DDI extraction method based on syntax CNN. In the method, the syntax word embedding, the position and part of speech features are introduced to extend the embedding of each word, and the auto-encoder is introduced to encode the traditional bag-of-words feature as the dense real value vector. The results of the CNN-based methods validate that CNNs have a great potential on DDIE tasks, however attention mechanisms can improve the performance of CNN. It emphasizes the important words in the sentence of a target-entity pair. It can automatically obtain the important feature of each channel by learning and utilize the important feature to enhance the classification ability and suppress the unimportant features to the current tasks. It shows the obvious advantages in DDIE. Asada et al. [13] proposed a DDIE method based on CNN with attention mechanism and evaluated it on the DDIExtraction-2013 task. Zheng et al. [14] proposed an effective approach to classify DDIs from the literature by combining recursive neural network (RNN) and long short-term memory network (LSTM) with attention mechanism. The method performs better with respect to recognizing not only close-range but also long-range patterns among words, especially for long, complex and compound sentences. Quan et al. [15] proposed a multi-channel CNN model to extract multiple semantic representations for DDIE, and the F1-score reached 70.2% on the DDIExtraction2013 corpus. Xu et al. [16] presented a DDIE method through full attention mechanism which can combine the user generated content information with contextual information and conducted a series of experiments on the DDIExtraction2013 dataset to evaluate their method. Yi et al. [17] proposed a RNN model with multiple attention layers for DDIE, and tested the model on the SemEval DDIExtraction2013 dataset. It is reported that the dependency information is useful in NLP task, and has been applied to DDIE. Liu et al. [18] proposed a dependency-based CNN (DCNN) for DDIE and found that the errors from dependency parsers are propagated into DCNN and affect the performance of DCNN.
To reduce error propagation, they designed a simple rule to combine CNN with DCNN to extract DDIs in long distances as most dependency parsers work well for short sentences but bad for long sentences. Zhang et al. [19] presented a DDIE method based on hierarchical recurrent neural networks (RNNs), shortest dependency path (SDP) and sentence sequence for extraction task and introduced the embedding attention mechanism to identify and enhance keywords for the DDIE task. In the method, the sentence sequence is divided into three subsequences, the bottom RNNs model is employed to learn the features of the subsequences and SDP, and the top RNNs model is employed to learn the features of both sentence sequence and SDP.
Gated Recurrent Unit (GRU) is a kind of deep learning using graph theory to learn temporal changes in a sequence, recently has been widely used in NLP [20]. Capsule network can improve the feature representation performance of CNNs and RNNs. It has achieved good results in the image field and the task of natural language processing [21]. It is able to capture the intrinsic spatial part-whole relationship constituting domain invariant knowledge that bridges the knowledge gap between the source and target tasks. Zhao et al. [22] proposed a hybrid model for DDIE by combining bidirectional gated recurrent unit (Bi-GRU) and GCN, where Bi-GRU and GCN are used to automatically learn the sequential representation and syntactic graph representation, respectively. By extracting the SDP of the two drug entities and considering the advantage of Bi-GRU in capturing long-distance sequence information, the original statement is combined with the SDP information to obtain low-level sentence representations containing more sentence information. Aiming at the problem of long sentences and complex sentence structure in medical texts, inspired by the advantage of Bi-GRU and Capsule networks in natural language processing [22], a DDIE method is proposed based on BiGCN and Capsule networks. Compared with most of the deep learning based DDIE approaches, the proposed method does not heavily rely on the quality of input instance representation and does not require any linguistic knowledge. The main contributions are as follows:
A DDIE method is proposed.
It is validated that the combination of Bi-GRU and Capsule networks does not require any linguistic knowledge to achieve state-of-the-art performance.
A lot of extensive experiments on a widely used dataset are conducted to show reasonable performance of capsule networks.
The rest of paper is arranged as follows. Section 2 introduces the related works, including Bi-GRU and capsule networks. The novel model is described in detail in Section 3. The experiments and results are presented in Section 4. Section 5 summarized the paper and points out the future works.
RELATED WORKS
Bidirectional Gated Recurrent Unit
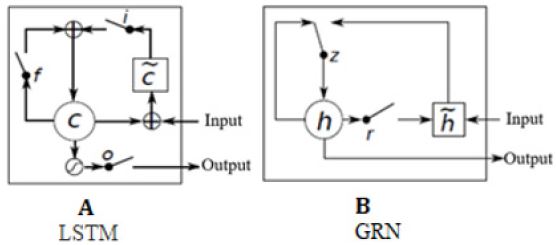
GRU is a variant of LSTM. Different from LSTM, it combines the forget gate and the input gate as a single update gate and adds the cell state and hidden state with other changes. It is simpler than the existing LSTM model and is a very popular variant, as shown in Figure 1. Bi-GRU, similar to bidirectional LSTM (Bi- LSTM), aims to learn the features from a sentence sequence whose outputs are later appended by GCN for DDIE. Its computations are divided into forward and reverse sequence information transmissions. Given a sentence, X= (x1, x2,..., xn), xt E Rk x denotes the concatenating vector of the current word and position, and the input and output of GRN is described as follows,
where 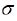 is a sigmoid activation function
is a sigmoid activation function 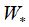 and
and 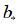 are the weight matrix and bias vector, respectively, xt is the input word vector at time step t and ht is the hidden state of the current time step t, is an element-wise product. Suppose the output of the forward GRU and backward GRU are
are the weight matrix and bias vector, respectively, xt is the input word vector at time step t and ht is the hidden state of the current time step t, is an element-wise product. Suppose the output of the forward GRU and backward GRU are  and
and 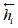 then the Bi-GRU output is denoted as
then the Bi-GRU output is denoted as
Capsule Network
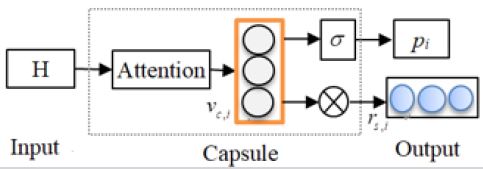
Capsule is a vector that can contain any number of values, representing a feature of the object that needs to be recognized at the moment. Each value of the convolutional layer of traditional CNN is the result of the convolution operation completed by a region of the previous layer and the convolution kernel, i.e., the linear weighted sum. It has only one value, so it is a scalar. Each value of the capsule network is a vector to detect the presence of features, which can not only represent the features of the object, but also include the orientation, position, direction and state of the object. The length of capsules reflects the probability of the presence of different features and the direction of capsules reflects the detailed properties of the features. Information between the layers is transmitted via a dynamic routing mechanism.
A capsule contains three modules: representation module,
probability module and reconstruction module, where representation module uses attention mechanism to build the
capsule representation 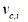 probability module uses sigmoid
function to predict the probability
probability module uses sigmoid
function to predict the probability 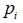 of the capsule active state, and
reconstruction module is developed from the capsule representation
and its state probability by multiplying
of the capsule active state, and
reconstruction module is developed from the capsule representation
and its state probability by multiplying 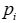 and
and 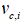 i.e.
i.e.  .
.
The capsule representation matches its attribute, and the state
of one capsule is corresponding to the input instance. Then, the
probability module based on the capsule representation will
be the largest if the capsule sentiment fits the input instance.
Reconstruction module is able to stand for the input instance
representation if its state is ‘active’ (Figure 2). The parameters
of capsule network are learned based on the aforementioned
objectives, i.e., maximizing the state probability of capsule selected
by ground truth sentiment, and minimizing the state probability of
another capsule. In testing, a capsule state will be active if 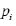 is the
largest among all capsules.
is the
largest among all capsules.
COMBINING BI-GRN AND FOR CAPSULE NETWORK FOR DDIE
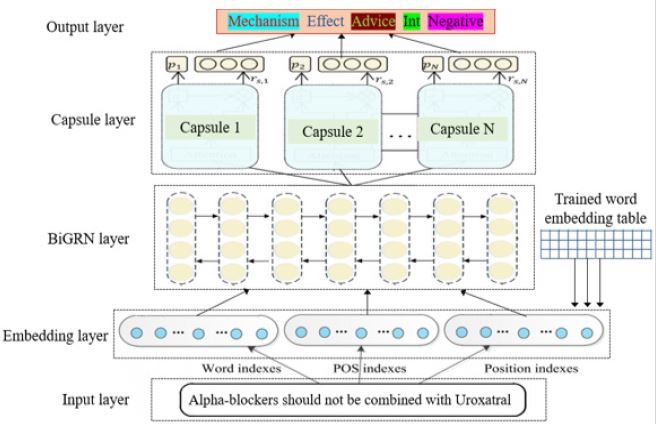
In this Section, we introduce the DDIE method based on Bi- GRN and for capsule network. Its architecture is shown in Figure 3, consisting of five layers: (1) In the input layer, input three kinds of information into the model, i.e., word, part of speech (POS) and relative distances between a word and each candidate drug in an input sentence; (2) In the embedding layer, look up the pre-trained word embedding vector table to encode the above input into realvalued vectors, called embedding vectors; (3) In the input attention layer, to further process the corresponding type of embedding features of a sentence into specific sequential data, and to learn the high-level syntactic meaning of the whole sentence and pass outputs at the last time step to the next layer; (4) In Capsule layer, calculate the similarity matrices to generate the prediction vector from a child capsule i to its parent capsule j; (5) In output layer, choose DDIs with top two probability meanwhile bigger than the threshold (empirically set the threshold 0.7) to perform DDI classification. The main layers are described in detail as follows.
The DDI corpus contains thousands of XML files, where each
is constructed by several records. For a sentence containing n
drugs, there are 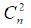 drug pairs. In DDIE, the set of text sentences
for the single drug pair or multiple drug pairs (maximum two drug
pairs in this paper) is denoted by . Suppose there are E predefined
DDIs (including Mechanism, Effect, Advice, Int, and Negative) to
extract. Formally, for each DDI r, the prediction type is denoted by
X = {x1, x2, ..., xn}.
drug pairs. In DDIE, the set of text sentences
for the single drug pair or multiple drug pairs (maximum two drug
pairs in this paper) is denoted by . Suppose there are E predefined
DDIs (including Mechanism, Effect, Advice, Int, and Negative) to
extract. Formally, for each DDI r, the prediction type is denoted by
X = {x1, x2, ..., xn}.
Suppose there are E predefined DDIs (including Mechanism, Effect, Advice, Int, and Negative) to extract. Formally, for each DDI r, the prediction type is denoted by P(r x1, x2, ..., xn).
Input Representation: For each sentence xi, the pretrained word embedding is employed to project each word token to the dw- dimensional space, each drug is represented as three features: the word itself, part of speech and position. The position features are used as the combinations of the relative distances from the current word to M drugs and encode these distances in Mdp -dimensional vectors, the POS feature is obtained by using the Stanford Parser to distinguish its semantic meaning [24].
Embedding: Each drug in the input sentence is mapped to a
real-valued vector representation using an embedding matrix
that is initialized with pre-trained embedding. Suppose 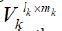 is the embedding table for the k-th feature group, mk is the
dimensionality of the embedding vector, and lk is the number of
features in the embedding table of a feature. Each embedding table
is initialized either by a random process.
is the embedding table for the k-th feature group, mk is the
dimensionality of the embedding vector, and lk is the number of
features in the embedding table of a feature. Each embedding table
is initialized either by a random process.
GRU: The hidden state ht in GRU denotes the representation of
position t while encoding the preceding contexts of the position by
Eq. (2), the Bi-GRU output is denoted as 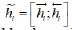 , where
, where 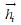 and
and 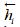 are the output of the forward GRU and backward GRU. Then the
instance representation, vs, is the average of the hidden vectors
obtained from GRU,
are the output of the forward GRU and backward GRU. Then the
instance representation, vs, is the average of the hidden vectors
obtained from GRU,
Where Ns is the length of instance, e.g., number of words in a given sentence.
Capsule: Given the hidden vectors encoded by GRU, the attention mechanism is used to construct capsule representation with a single parameter inside a capsule, as follows,
Where ht is the representation of word at position t, i.e., the hidden vector from GRU, and wt,i is the parameter of capsule i for the attention layer, and at,i is the attention important score for each position.
In Eq. (4), at,i can be obtained by multiplying the
representations with the weight matrix, and then normalizing to a
probability distribution over the words 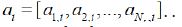 . .
The capsule representation vector vc,i is a weighted summation over all the positions using the attention importance scores as
weights.
. .
The capsule representation vector vc,i is a weighted summation over all the positions using the attention importance scores as
weights.
After getting the capsule representation vector vc,i, the active state probability pi calculates by
Where Wp,i and bp,i are the learning parameters for the active
probability of the current capsule i, 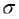 is the sigmoid function.
is the sigmoid function.
The parameters Wp,i and bp,i are learned through maximizing the state probability of capsule selected by ground truth sentiment and minimizing the state probability of another capsule. In testing, a capsule’s state will be active if pi is the largest among all capsules. The input instance is reconstructed by multiplying vc,t and probability pi,
The probability module based on the capsule representation is the largest if the capsule sentiment fit the input instance, while the reconstruction module is developed from the capsule representation and its state probability, so the reconstruction representation can stand for the input instance representation if its state is active.
Finally, given drug pair (e1, e2), the pretrained embedding of drugs and DDIs is adopted and calculated as follows,
Where t, h are the embedding of drugs e1, e2 respectively, rk is the relation embedding.
DDI with the closest embedding to the drug embedding difference is the classified type.
From the above analysis, the steps of the GRUTC based DDIE methods are listed as follows,
1) Data pre-processing layer: pre-processing the data,
mapping the data to the corresponding word vector and part-ofspeech
label vector after cleaning, spatial mapping, vector matrix,
and then input to the BI-GRN layer;
2) Bi-GRN layer: The processed data is input into the BI-GRN
network for feature extraction.
3) Capsule layer: The primary Capsule module integrates the
word feature and position feature into the primary Capsule, carries
out the dynamic routing algorithm based on attention mechanism
between capsules, and calculates and inputs the DDI classification
layer through the feature clustering layer.
4) DDI classification layer: DDI classification was carried
out based on the probability obtained with Capsule to get the final
result.
EXPERIMENTS AND RESULTS
In this Section, the proposed DDIE method is evaluated on the DDI Extraction 2013 corpus database, and compared with a traditional DDIE approach and three deep learning methods: linear kernel approach (FLK) [9], CNN [11], syntax CNN (SCNN) [12] and LSTM [25]. Three official evaluation metrics F-Score (F), Precision (P) and Recall (R) are adopted to evaluate the effectiveness of the proposed model:
Where True Positive (TP) is number of DDIs when the candidate DDI matches the real DDI, False Positive (FP) is number of DDIs when the candidate DDI does not match the real DDI, and the number of False Negative (FN) is calculated by calculating the DDI not detected by the model.
Data
DDIExtraction 2013 corpus dataset was constructed from January to June 2013 and attracted much attention with a total of 14 teams from 7 different countries, where 6 teams participated in the drug name recognition tasks, while 8 teams participated in the DDI extraction tasks. The database consists of 1017 XML documents, including 784 texts selected from the Drug Bank database and 233 abstracts regarding to DDIs selected from the MEDLINE database [25]. It can be unloaded from http://www. cs.york.ac.uk/semeval-2013/task9/.
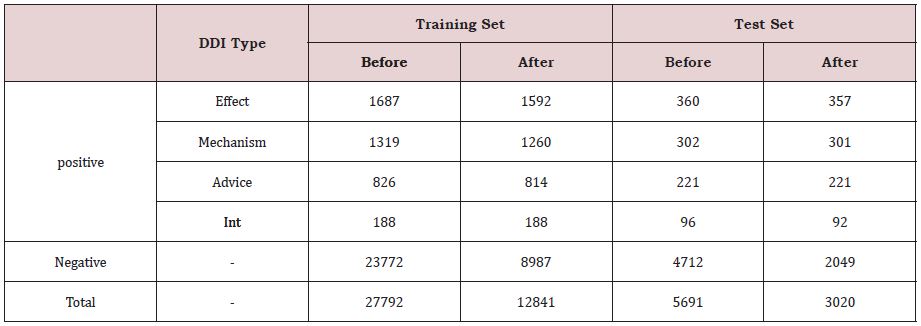
All drug pairs in each sentence were annotated manually as either no DDI or true DDI. The database is split into training and test instances provided by the sentences. It is a multi-classification task, annotated with five annotation types, namely Negative, Mechanism, Effect, Advise, and Int. Their statistics is shown in Table 1.
From Table 1, it is seen that the database is extremely unbalanced. That is to say, there are large negative samples. Many DDIE methods often suffer from the imbalanced class distribution problem, which will significantly affect their classification performance. It is known that filtering out the negative instances can improve the DDIE ability. Therefore, we filter out the negative instances on the entire dataset based on the predesigned rules [12], the filtered statistics are also shown in Table 1.
According to the drug entity in the original statement, the drug pair of interaction is generated. For generalizing our model, the drug blind treatment is conducted, namely, the two drugs in pair are respectively replaced with “DRUG_1” and “DRUG_2” in turn by the filtering rules in [12], and all the other drugs in the same sentence are replaced by “DRUG_N” [11-15], where “DRUG_1” and “DRUG_2” represent the drug entity to judge the interaction relationship, and “DRUG_N” represents the unrelated drug entity.
Experimental Set
In the experiment, the DDIE experiments are conducted on Keras and TensorFlow1.7.0 framework, including LSTM, Ubuntu18.04LTS as the operating system, 32G memory, Intel Core i5-4200U CPU @2.30 GHz, GPU GEFORCE GTX 1080ti, Ubuntu14.0, the embedding of word and position is initialized randomly, and their embedding vector dimensions are set as 200 and 30, respectively, the hidden state’s dimension is 230, the probability of dropout is 0.5, other hyper-parameters which are not shown here are set to TensorFlow default values. The word embedding is initialized by pre-trained word vectors using GloVe [26], while other parameters are initialized randomly. During each training step, a mini-batch (size is set 60 in our implementation) of sentences is selected from the training set. The gradient of objective function is adopted to update the parameters. We record the result every 100 step and classify all the DDIs of the sentence of the test set.
RESULTS
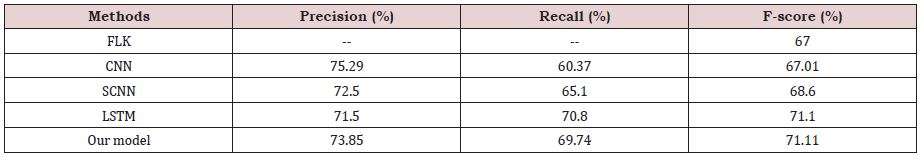
The overall results of the proposed method and 5 comparative methods on the test set are shown in Table 2.
From Table 2, it is found that the traditional DDI extraction method is poor, because its F1-score mainly relies on a large number of the handcraft features to improve its performance, resulting in high system cost and low generalization ability, while the deep learning models can achieve good results without using any handcraft features, and the F1-score of the proposed method is greatly improved. The proposed method outperforms other approaches, because the combination of the shortest dependent path information can better enrich the semantic information of the sentence, which is better than the use of the original sentence information alone, and the ability of the capsule network to dynamically utilize the high-level information can significantly improve the effect of the model. The results indicate that dynamic routing may be more effective than the max-pooling operator in CNN, which essentially detects whether a feature is present in any position of the text or not but loses spatial feature information.
CONCLUSION
DDIE has become a vital part of public health safety and extracting DDIs using text mining techniques from biomedical literature has received great attentions. However, this research is still at an early stage and its performance has much room to improve. In this paper, we proposed a novel DDIE approach based on Bi-GRU and capsule network. In the method, Bi-GRU can capture long distance information, capsule network can obtain the highlevel information, and the shortest dependent path information can be used to enrich the statement information. The experimental results validate that this method is effective in extracting DDIs from complex medical texts. In the future work, we try to add attention mechanism to the original sentence information and the shortest dependency information, so as to improve the effect of the model. In view of the phenomenon that there are many negative cases in the common dataset, data enhancement and other operations can be considered in future work to balance the quantitative difference between different types of data.
ACKNOWLEDGMENT
This work is supported by the National Natural Science Foundation of China (No. 62072378).
REFERENCES
- Hakkarainen KM, Sundell KA, Petzold M (2012) Methods for assessing the preventability of adverse drug events. Drug Saf 35(2): 105-126.
- Percha B, Altman RB (2013) Informatics confronts drug-drug interactions. Trends Pharmacol Sci 34(3): 178-184.
- Midtvedt T (2007) 25 Penicillins, cephalosporins, other beta-lactam antibiotics and tetracyclines. Side Effects of Drugs Annual 29: 244-252.
- Ayvaz S, Horn J, Hassanzadeh O (2015) Toward a complete dataset of drug-drug interaction information from publicly available sources. Journal of Biomedical Informatics 55: 206-217.
- Óscar EJ, Cristina NP, Francisca GR (2017) A study of incidence and clinical characteristics of adverse drug reactions in hospitalized patients. Rev Esp Salud Publica 91: e201712050.
- Thiesen S, Conroy EJ, Bellis JR (2013) Incidence, characteristics and risk factors of adverse drug reactions in hospitalized children-a prospective observational cohort study of 6,601 admissions. BMC Med 11(1): 1-10.
- Vilar S, Uriarte E, Santana L (2014) Similarity-based modeling in largescale prediction of drug-drug interactions. Nature Protocols 9(9): 2147- 2163.
- Mezaache S, Comont T, Germain J (2015) Incidence of adverse drug reactions related to immune thrombocytopenia drugs. A prospective cohort studies. Blood 126(23): 1056-1056.
- Kim S, Liu H, Yeganova L (2015) Extracting drug-drug interactions from literature using a rich feature-based linear kernel approach. Journal of Biomedical Informatics (55): 23-30.
- Zhang W, Yue X, Liu F (2017) A unified frame of predicting side effects of drugs by using linear neighborhood similarity. BMC Systems Biology 11(S6): 101.
- Shengyu L, Buzhou T, Qingcai C (2016) Drug-drug interaction extraction via convolutional neural networks. Comput Math Methods Med 2016: 6918381.
- Zhao Z, Yang Z, Luo L, Lin H (2016) Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics 32(22): 3444-3453.
- Asada M, Miwa M, Sasaki Y (2017) Extracting drug-drug interactions with attention cnns. proceedings of the BioNLP 2017 workshop, Vancouver, Canada 4: 9-18.
- Zheng W, Lin H, Luo L (2017) An attention-based effective neural model for drug-drug interactions extraction. BMC Bioinformatics 18(1): 445.
- Quan C, Lei H, Sun X (2016) Multichannel convolutional neural network for biological relation extraction. BioMed Research International 2016(2-1): 1-10.
- Xu B, Shi X, Yin Y (2019) Incorporating user generated content for drug drug interaction extraction based on full attention mechanism. IEEE Transactions on NanoBioence, 2019, 18(3):360-367.
- Zibo Yi, Shasha Li, Jie Yu (2017) Drug-drug interaction extraction via recurrent neural network with multiple attention layers. Advanced Data Mining and Applications 2017: 554-566.
- Liu S, Chen K, Chen Q (2016) Dependency-based convolutional neural network for drug-drug interaction extraction. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2016: 1074- 1080.
- Zhang Y, Zheng W, Lin H (2018) Drug-drug interaction extraction via hierarchical RNNs on sequence and shortest dependency paths. Bioinformatics 34(5): 828-835.
- Pil-SooKim, Dong-GyuLee, Seong-WhanLee (2018) Discriminative context learning with gated recurrent unit for group activity recognition. Pattern Recognition 76: 149-161.
- Min Y, Wei Z, Lei C, Qiang Q, Zhou Z, et al. (2019) Investigating the transferring capability of capsule networks for text classification. Neural Networks 118: 147-261.
- Di Z, Jian W, Hongfei L (2019) Extracting drug-drug interactions with hybrid bidirectional gated recurrent unit and graph convolutional network. Journal of Biomedical Informatics 99: 103295-103295.
- Haridas P, Chennupati G, Santhi N (2020) Code characterization with graph convolutions and capsule networks. IEEE Access (99):1-1.
- De Marneffe MC, MacCartney B, Manning CD (2006) Generating typed dependency parses from phrase structure parses. Proceedings of LREC 2006: 449-454.
- Kumar SS, Ashish A (2017) Drug-drug interaction extraction from biomedical text using long, short term memory network. Journal of Biomedical Informatics 86: 15-24.
- Isabel SB, Paloma M, María HZ (2014) Lessons learnt from the DDIExtraction-2013 shared task. Journal of Biomedical Informatics 51: 152-164.
Article Type
Research Article
Publication history
Received date: December 22, 2020
Published date: January 07, 2021
Address for correspondence
Wenzhun Huang, College of Information Engineering, Xijing University, China
Copyright
©2021 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Shanwen Z,Wenzhun H, Yun Z. Drug-Drug Interaction Extraction Based on Bidirectional Gated Recurrent Unit networks and Capsule Networks. 2021- 3(1) OAJBS. ID.000247.