Development of High Throughput Screening to Discover SARS-CoV-2 Main Protease Inhibitors with NanoBit Complementation
ABSTRACT
SARS-CoV-2 has become a serious global public health problem and there is lack of effective drugs to fight it. Main protease (Mpro) of SARS-CoV-2 is an important antiviral drug target. We aim to establish a high-throughput screening (HTS) assay to identify inhibitors of SARS- CoV-2 Mpro. A Mpro NanoBiT assay for HTS was developed by using NanoBiT complementary strategy. Mpro peptide substrate was flanked by subunits of Nano luciferase (NanoLuc), SmBiT and LgBiT respectively at the amino and carboxyl termini to obtain the structure Mpro cleavage site-NanoBiT. Mpro cleavage site-NanoBiT showed strong NanoLuc activity, which was modulated with Mpro peptide digestion, indicating the inhibitory effect of compounds on Mpro could be read out with NanoLuc activity. We optimized the experiment conditions including the quantity of Mpro and Mpro cleavage site-NanoBiT, DMSO tolerance, enzyme digestion time, and plate readout time after adding NanoLuc substrate. We further evaluated with two Mpro reference inhibitors, GC376 and Mpro inhibitor 11a, and found the assay was suitable for HTS of Mpro inhibitors. The miniaturized 384-well plate format assay was further used to screen 6,400 compounds from FDA-approved Drug Library and Natural Bioactive Compound Library. Control experiments showed a significant separation of relative activity of the negative and positive controls leading to an acceptable HTS Z’ value of 0.67, indicating satisfactory assay performance. Our results demonstrated that our established Mpro NanoBiT assay by inserting protease substrate peptides between SmBiT and LgBiT of NanoBiT could be applied for discovery of inhibitors of different proteases.
KEYWORDS
SARS-CoV-2; Mpro; High-throughput screening; NanoBiT complementation; Protease inhibitor
INTRODUCTION
The occurrence and rapid spreading of the highly pathogenic coronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have become a serious global public health emergency [1,2]. In severe cases, patients rapidly progress to acute respiratory distress syndrome, septic shock, difficulty to correct metabolic acidosis, and coagulation dysfunction [3]. The estimated mortality rate is 3%-5% [4,5]. However, there are few specific antiviral drugs to fight against this virus [6,7]. It is thus pivotal and urgent to search for effective therapeutic agents to tackle SARS-CoV-2. SARS-CoV-2 binds to the host receptor and enters the cells through fusion with the cell membrane. The virus nucleic acid is then released into the cytoplasm, and the viral proteins are translated by host ribosomes [8]. The main protease (Mpro), or 3C-like protease, of the virus hydrolyzes polyproteins to produce functional proteins, such as RNA-dependent RNA polymerase, which is essential for virus replication and gene transcription [9]. Furthermore, Mpro sequence is highly conserved among SARS-CoV, MERS-CoV and SARS-CoV-2 [8,10], and is responsible for virusinduced apoptosis signals that may lead to the death of surrounding uninfected cells [11]. It has been proposed that targeting Mpro with antiviral drugs may not only inhibit virus replication, but also suppress the signal cascade in infected cells to protect uninfected cells. Therefore, Mpro is a promising antiviral drug target [12,13].
Up to now, three strategies have been applied to explore the discovery of Mpro inhibitors. The first strategy used computeraided drug design according to the crystal structure of SARS-CoV-2 Mpro [14]. Fluorescence resonance energy transfer (FRET) assays were recently developed by pairing donors and receptors into Mpro peptide substrate to assess the cleavage rate of the substrate [15,16]. Moreover, a GFP-split-Mpro high-throughput screening system was established with split-GFP complementation, in which GFP-based reporters became fluorescent upon cleavage by SARSCoV- 2 protease Mpro [12]. All three methods have been used to screen compound libraries to discover potential SARS-CoV-2 protease Mpro inhibitors by HTS. However, the complexity of these systems has limited their wide application for the identification and validation of effective Mpro inhibitors for their rapid entry into clinical trials. It is therefore important to develop alternative and feasible assays to screen SARS-CoV-2 protease Mpro inhibitors by HTS.
In the present study, we developed and optimized Mpro-NanoBiT platform for HTS to identify inhibitors against SARS-CoV-2 by inserting Mpro substrate between SmBiT and LgBiT of NanoBiT. In this system, the compound inhibitory activity to Mpro could be reported with NanoLuc activity. Our results suggest that this system is a practical and reliable platform for HTS to identify Mpro inhibitors against SARS-CoV-2 as well as inhibitors of other virus proteases.
MATERIALS AND METHODS
Compound Libraries
The compound libraries used in this study for HTS of SARSCoV- 2 Mpro inhibitors are FDA approved Drug Library (Targetmol; MA, USA) and Natural Bioactive Compound Library (Targetmol; MA, USA). The compounds were dissolved in DMSO at 10mM and stored at -80 °C.
Construction of pLVX-Mpro Cleavage Site-NanoBiT-hyg
The DNA fragments encoding signal peptide for secretion, SmBiT, Mpro cleavage site, LgBiT and SNAP were synthesized from Genecreate (Genecreate; Wuhan, China) and inserted into pLVXIRES- hyg (Takara; CA, USA) between sites of XhoI and XbaI (NEB; MA, USA). SmBiT, LgBiT and SNAP sequences were from pBiT2.1-N TK/SmBiT, pBiT1.1- C TK/LgBiT vectors (Promega; WI, USA) and pSNAPf vector (NEB, MA, USA). Plasmids were amplified in Stbl3 competent cells (ZOMANBIO; Beijing, China) and purified with an endotoxin-free plasmid medium extraction kit (TIANGEN; Beijing, China). Prepared plasmids were quantified, aliquoted and stored at -20 °C for transfection.
Cell Culture, Transfection and Production of Lentivirus
HEK 293T cells (BBI; Shanghai, China) were maintained in DMEM high glucose medium (Corning; VA, USA) containing 10% FBS (Fetal bovine serum, Gibco; NY, USA), 1% penicillin and streptomycin (Hyclone; UT, USA), and cultured at 37 °C in a 5% CO2 incubator. When the cells reached 80-90% confluence in 100mm cell culture dishes, the medium was replaced with Opti-MEM (Gibco; NY, USA) at 5mL per well. The cells were transfected with 2μg pLVX-Mpro cleavage site-NanoBiT-hyg plus help vectors with LipofectamineTM 2000 (Invitrogen; CA, USA) according to the manufacturer’s instruction. The supernatant medium was collected between 48 and 72 hours and virus titer (multiplicity of infection, MOI) was determined with QuickTiter™ Lentivirus Titer Kit (Cell Biolabs Inc; CA, USA). After collection, the virus was filtered with a 0.45μm filter and centrifuged at 4 °C with 2000g for 120min in a 40mL ultracentrifuge tube. The virus precipitate was resuspended with 500μl of fresh medium, aliquoted and stored under -80 °C.
Immunoblotting of Mpro Cleavage Site-NanoBiT
Mpro cleavage site-NanoBiT in 15μL of supernatant from 2μg pLVX-Mpro cleavage site- NanoBiT-hyg transfection in a 35mm petri dish were separated on sodium dodecyl sulfate- polyacrylamide gel electrophoresis gels (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (PVDF, Millipore; MA, USA). The membranes were blocked with 5% BSA in tris buffered saline with Tween-20 (TBST) and incubated with a His-Tag antibody (Yeasen; Shanghai, China). Then the membranes were incubated with IRDye 800CW Goat Anti-Rabbit IgG (H + L) (LI-COR; NE, USA) for 2 hours at room temperature and the signals were detected with Odyssey® CLx Imaging System (LI-COR; NE, USA). Specific bands were evaluated by the predicted molecular size.
Production of Mpro Cleavage Site-NanoBiT
The packaged viruses were taken out from -80 °C freezer and thawed on ice. HEK 293 cells (Obio; Shanghai, China) were maintained in DMEM high glucose medium containing 10% FBS and 1% penicillin/streptomycin and cultured at 37 °C in a 5% CO2 incubator.
When the cells were grown to 80-90% confluence in 150mm cell culture dishes, the supernatant medium was removed. Five MOI of viruses (5-10mL) were transduced into HEK 293 cells, and fresh medium was replaced at 4-6 hours post transduction. The supernatant medium was collected 48 and 72 hours after transduction, and stored at -20 °C.
Purification of Mpro Cleavage Site-NanoBiT
The Mpro cleavage site-NanoBiT was purified with a His-tag kit (Abbkine; CA, USA). The supernatant was mixed with 1mL of preequilibrated (buffer A) Ni-NTA-beads and incubated on a rolling shaker for 30min at 4 °C. Subsequently, the Ni-NTA-beads were washed with buffer A, containing increasing concentrations of imidazole (10mM, 20mM, 50mM, pH 7.6; 10-20mL buffer per step). The protein was eluted by multiple elution steps, each with 1 mL of buffer B (20 mM Tris, 200 mM NaCl, 500mM imidazole). Elution fractions were analyzed by OD measurement and SDS-PAGE. Fractions containing SARS- CoV-2 Mpro cleavage site-NanoBiT were concentrated, and the buffer was exchanged with buffer C (20mM Tris, 200mM NaCl, 1mM DTT, 1mM EDTA, pH 7.6) using Amicon centrifugation filters (MERCK; Darmstadt, Germany). The proteins were further purified by size-exclusion chromatography using an S200 column with buffer C as running buffer.
Fractions with pure protein were concentrated and mixed with 50% sterile glycerol, flash frozen in liquid nitrogen at aliquots of 1mg/mL and stored at -80 °C until further use.
ELISA to Measure purified Mpro concentration
A mouse monoclonal anti-DYKDDDDK (Flag Tag epitope) antibody (SinoBiological; Beijing, China) was coated on a microtiter plate, and then incubated with different concentrations of Mpro cleavage site-NanoBiT (two-fold dilution, 200pg-0.1μg/mL) at 37 °C for 1 hour. After washing with phosphate buffered saline with Tween-20 (PBST), the plate was incubated with anti His- Tag antibody (4μg/mL) for 1 hour, followed by incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit antibody (SinoBiological; Beijing, China) (dilution ratio 1:6000) for 1 hour. After staining, the OD values were measured at 450nm using a fullwavelength microplate reader (BioTek Instruments, Inc.; VT, USA).
NanoLuc Activity Test of Mpro Cleavage Site-NanoBiT
In a 384-well plate (Corning; VA, USA), the purified Mpro cleavage site-NanoBiT (100ng/mL) was diluted to 20-fold, 30-fold, 40-fold, 50-fold, and 60-fold gradients with DMEM to test NanoLuc activity. The dilution was conducted with manual pipettes. The final volume of dilution for each well was around 10μL. Then 15μL of Nano- Glo® HiBiT Extracellular Detection System luminescence substrate (Promega; WI, USA) was added with liquid handler, Apricot Designs S-Pipette S2 (Apricot Designs; CA, USA). After addition of samples into the wells, the plates were mixed and centrifuged, followed by reading with Tecan Spark multifunctional microplate reader (Tecan; Männedorf, Switzerland).
Optimization of Mpro Concentration
Mpro (Beyotime; Shanghai, China) was diluted to 0.25μg/μL. Ten μL of 50-fold diluted Mpro cleavage site-NanoBiT was split into each well of a 384-well plate for digestion, followed by addition of different dosages of Mpro (0, 0.25, 0.5, 1, 1.25). All wells were filled up to 25μL with DMEM. After addition 25μL of Nano luminescence substrate to each well, the plates were mixed, centrifuged, and read with a microplate reader.
Optimization of DMSO Concentration
Ten μL 50-fold diluted Mpro cleavage site-NanoBiT was added into each well of a 384- well plate. DMEM (15μL) with different concentrations of DMSO (Nacalai Tesque; Kyoto, Japan) was added into the wells to make 25μL volume with 0%, 0.25%, 0.5%, and 1% of DMSO. Finally, 25μL of Nano luminescence substrate was added. At 1min, 5min, 10min, and 15min after adding samples to the wells, the plates were mixed, centrifuged and read with a microplate reader.
Screening Process of Mpro-NanoBiT Assay
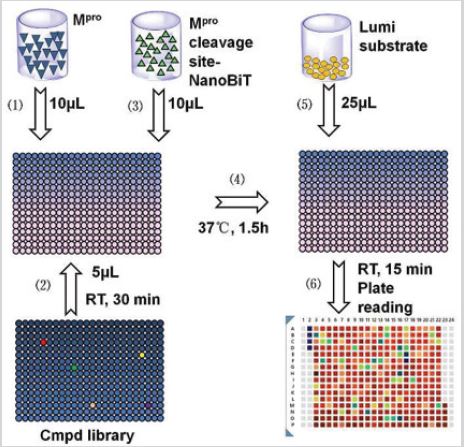
Mpro was diluted at 1 unit per 10μL with DMEM plus 2.5mM DTT (Thermo Fisher Scientific; MA, USA) and split into 384-well plates (10μL/well) with liquid handler, Apricot Designs S-Pipette S2. Five μL of 100μM of each compound from the compound libraries was added into the plates. After mixing, the plates were centrifuged to collect the solution to the bottom of wells.
The plates were stored at room temperature for 30 minutes to allow the compounds to fully bind to Mpro. Wells of A2-D2 were filled with 0.5% DMSO; Wells of M23-P23 were filled with GC376 to a final concentration of 300nM. Then 10μL of diluted Mpro cleavage site-NanoBiT was added, mixed and centrifuged. All plates were placed at 37 °C for 1.5 hours and Nano luminescence substrate (25μL) was added into each well. After mixing and centrifuge, the plates were incubated at room temperature for an additional 10- 20min and subjected to read with a microplate reader. All signals were picked up with integration time of 100 microseconds.
Statistics
GraphPad Prism Software version 7.0 (GraphPad Software Inc.; CA, USA) was used for data analysis and graph plotting.
The Z’ factor was calculated as follows: Z’ = 1-((3σc++3σc-) / (|μc+-μc-|)), where σ represents standard deviation; μ represents mean; c+ represents positive control; and c- represents negative control.
RESULTS
Design of Mpro-NanoBiT Assay
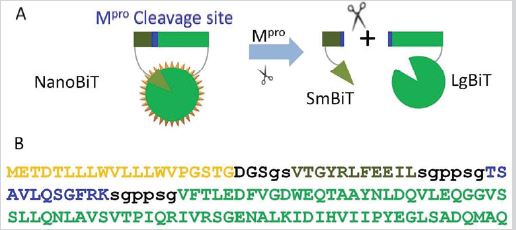
The principle of NanoBiT is to fuse two luciferase subunits LgBiT and low affinity SmBiT to the termini of two proteins which can interact with each other leading to the proximity and formation of functional luciferase by LgBiT and SmBiT. We hypothesize that, when the two low affinity subunits LgBiT and SmBiT are linked together with a short peptide containing the main protease cleavage site, this structure should have strong NanoLuc activity; in contrast, when the short peptide is cleaved by Mpro, the two subunits will be separated and not be able to form a functional NanoLuc (Figure 1A). To test this hypothesis, according to the NanoLuc crystal structure 7mjb [17], we first simulated a homolog model of protein structure of Mpro cleavage site-NanoBiT with online tool of Swiss Model (https://swissmodel.expasy.org/) (data not shown). In this structure, SmBiT and LgBiT were respectively flanked at the amino and carboxyl termini of Mpro cleavage site plus appropriate linkers. To facilitate the secretion of the expressed protein, the signal peptide of IgG kappa was used as the leading sequence. Histag, FLAG-tag and SNAP were attached to the carboxyl of LgBiT. The details and amino acid sequence of the whole structure are shown in Figure 1B. The SNAP tag is convenient to detect fusion protein expression.
Expression and NanoLuc Activity of Mpro Cleavage Site- NanoBiT
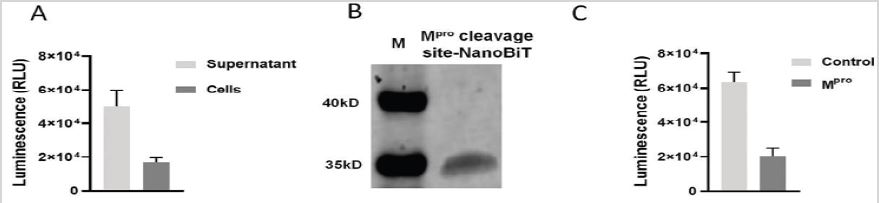
To determine whether the designed Mpro Cleavage Site-NanoBiT can be secreted into cell culture medium and has NanoLuc activity, the pLVX-Mpro cleavage site-NanoBiT-hyg was constructed and introduced into HEK 293T cells in 12-well plate. After 48 hours, the cell culture medium was collected as the supernatant, and the cells were trypsinized and washed twice with PBS to remove traces of secreted NanoLuc. NanoLuc activities of 25μL supernatant and 10,000 cells in 25μL DMEM with 10% FBS were measured. The results showed that Mpro cleavage site-NanoBiT was mainly secreted into the cell culture medium, with 50kRLU (relative luminescence unit) in supernatant while only 17k RLU in the cells (Figure 2A).
The luminescence from non-transfected cells and the corresponding supernatant showed background of 50 to 200RLU with Tecan Spike reader. Further immunoblotting of 15μL of supernatant with an anti His-Tag antibody detected a protein of about 35kD, close to the expected 34kD of Mpro cleavage site- NanoBiT (Figure 2B).
To determine whether the Mpro cleavage site inserted between the two subunits of NanoLuc is accessible to and can be cleaved by Mpro, 5μg of Mpro was added into 25μL supernatant in 50μL reaction system. After 2 hours of digestion, the luminescence was detected and found to be decreased from 63k RLU to 20k RLU (Figure 2C).
These results demonstrated that the insertion of Mpro cleavage site into NanoLuc between SmBiT and LgBiT did not disrupt the enzyme activity and can be accessed to and cleaved by Mpro, providing the basis for further development of the assay.
Optimization of Mpro-NanoBiT Assay
In order to meet the requirement for extensive optimization and evaluation of an HTS assay for screening thousands of compounds, a large amount of Mpro cleavage site-NanoBiT was produced with lentivirus transduced HEK 293 cells. Moreover, to eliminate the potential impact of the proteins in FBS on the activity of Mpro, Mpro cleavage site-NanoBiT was purified with a His-Tag purification kit and aliquoted for further characterization.
Firstly, it is necessary to optimize the dilution factor of Mpro cleavage site-NanoBiT to obtain the best luminescence signals. During screening drugs by HTS, the luminescence value may reach the upper limit of the detection instrument and fail to be detected when the value is too high; conversely, the inhibitory effect will not be read out when the value is too low.
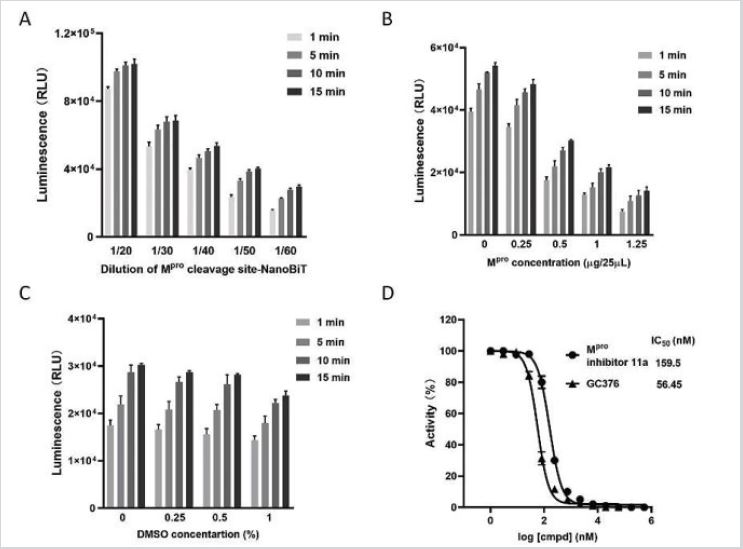
Moreover, the right concentrations of the substrate for Mpro are critical as a high Mpro substrate concentration can reduce the potencies (IC50s) of inhibitors determined in enzyme assays [18]. For these reasons, we determined the luminescence value using different concentrations of purified Mpro cleavage site-NanoBiT and found the luminescence values were among 40-120k RLU when Mpro cleavage site-NanoBiT was diluted in 20, 30, 40, 50, and 60 folds (Figure 3A). Accordingly, the dilution factor of Mpro cleavage site-NanoBiT was determined to be 50 times according to the luminescence value. NanoLuc Activity of Mpro Cleavage Site- NanoBiT were varied according to the batches of purification and storage time. For assays in this research, all experiments were done based on the absolute RLUs of NanoLuc Activity of Mpro Cleavage Site-NanoBiT.
Secondly, Mpro belongs to the category of cysteine proteases and its enzyme activity can be strongly modulated by DTT which prevents intramolecular or intermolecular disulfide bonds formed between cysteines in the proteins [19]. In our assays, all enzyme reactions contained 1mM DTT to reduce possible false positive hits. To optimize the concentration of Mpro, Mpro cleavage site-NanoBiT was diluted to 50-fold and added at 10μL/well, while Mpro was added with concentrations of 0, 0.25, 0.5, 1, 1.25μg/well in 384- well plates. The final volume was brought to 25μL with DMEM. After digestion at 37 °C for 1 hour, the NanoLuc signals were reduced by 60% to 20k RLU in the reaction containing of 1μg Mpro (Figure 3B). To balance the signals and reduce Mpro enzyme cost, 1μg Mpro for the system was determined.
Thirdly, since the compounds in the libraries to be screened were dissolved in DMSO, a right DMSO concentration in the reaction system should be optimized. Theoretically, a high concentration of DMSO facilitates compound distribution, but could inhibit the activity of the two enzymes in the reaction system: Mpro mediated digestion of Mpro cleavage site-NanoBiT and Mpro cleavage site- NanoBiT catalyzed luminescence production. To determine an appropriate DMSO concentration, 50-fold diluted Mpro cleavage site- NanoBiT plus 1μg Mpro were mixed with DMSO in the concentration of 0, 0.25, 0.5, 1% and split into 384-well plates. The results indicated that 0.5% DMSO would maintain the compounds well dissolved and did not apparently affect the activity of the enzymes (Figure 3C).
Finally, to validate the enzyme assay, two known SARS-CoV-2 Mpro inhibitors, GC376 [20] and 11a [21], were evaluated in 384- well format. In the presence of 60-fold diluted Mpro cleavage site- NanoBiT, 1μg of Mpro, 1mM DTT and 0.5% DMSO, the IC50s of GC376 and 11a were 56.45nM and 159.5nM, respectively (Figure 3D). These results demonstrated that our established Mpro-NanoBiT assay was suitable for HTS to discover Mpro inhibitors.
High Throughput Screening of FDA-Approved Drug Library and Natural Bioactive Compound Library with Mpro-NanoBiT Assay
After optimization of the assay critical parameters, the assay was miniaturized to fit 384-well plate format. In brief, the original compound libraries were purchased in a 384-well format at a concentration of 10mM. Daughter compound libraries were prepared as 100μM in 2.5% DMSO. Compounds were tested between columns 3 and 22. The detailed processes were described in Figure 4.
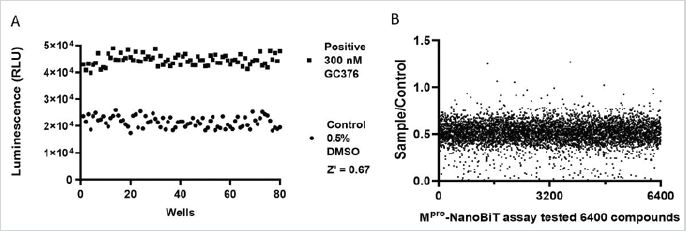
A total of 6,400 compounds (10 of 384-well plates of FDAapproved Drug Library and 10 of 384-well plates of Natural Bioactive Compound Library) were screened with our optimized Mpro-NanoBiT assay. For quality control, 4 wells of positive (300nM GC376) and negative (0.5% DMSO) controls were set in each plate. Control experiments showed a significant separation of relative activity of the negative and positive controls leading to an acceptable HTS Z’ value of 0.67, demonstrating signal consistency and satisfactory assay performance (Figure 5A). In the Mpro- NanoBiT assay, Mpro cleavage site-NanoBiT added into the wells showed about 50k RLU. After addition of Mpro and incubation at room temperature for 1.5 hours, the signals were decreased to 20k and 40k RLU respectively in the presence of 0.5% DMSO only. All data were normalized with negative controls in the same plates and the scatter plots were shown in Figure 5B. Compounds which have a value more than 0.5 were considered to have inhibitory effect.
DISSCUSION
Proteases play an important role in many signaling pathways and represent potential drug targets for diseases ranging from cardiovascular disorders to cancers, as well as many parasites and viruses [22,23]. There are many kinds of proteolytic enzymes, and their substrates are even more complicated. If the chemical FRET method is applied, each digested substrate must be labeled with FRET chromophores, which is inconvenience. Compared with the GFP-split assay, Mpro-NanoBiT assays have much wide windows to pick up signals. Thus, the NanoBiT complementation method developed here can also be adapted to screen inhibitors of other proteases.
The advantage of this protocol is that the assay is relatively simple, economical and has certain universal adaptability. Most laboratories can launch similar experiments to perform small batches of protease inhibitor activity analysis with the compounds in which they are interested. The assay can be miniaturized and used for high-throughput screening. After micro quantification, the amount of reagents used is further reduced, which will greatly reduce the cost of experiments. In addition, the use of the substrate to detect the activity of protease inhibitors has the advantages of sensitivity, speed, strong signal and controllable cost.
Generally, there were many false-positive hits in the primary HTS screening. In most cases, the primary hits are about 2-3 percent [24]. In our Mpro-NanoBiT assay, after filtering obvious artificial hits [25], if the cutoff was set as 60% inhibitory effect, there were 95(1.48%) hits, indicating a relative reasonable hit percentage. False negative and positive hits could be caused by many factors including but not limited to the following reasons. First, the compounds which bind to NanoLuc and inhibit NanoLuc activity may mimic Mpro activator phenotype. Second, the compounds which bind to Mpro cleavage site-NanoBiT and block Mpro access to the peptide substrate might also mimic Mpro activator phenotype. Third, the compounds which activate Nano-luciferase could mimic Mpro inhibitor phenotype. Thus, NanoLuc reporter assays, including ours, also have limitations. The compounds that are able to inhibit the NanoLuc activity could also cause false negative hits, and vice versa. Unlike firefly luciferase [26], there are no references of NanoLuc inhibitors available yet. This shortage could be avoided by parallel screening with a similar strategy. In the future, similar NanoBiT assays should be established with two constructs such as RBD-SmBiT and LgBiT- sACE2. Moreover, the same compound libraries should be counter screened to each other to filter possible negative or positive false hits. In addition, those selected compounds should be grouped according their chemotypes and further verified with different methods before testing in vivo.
CONCLUSION
In this study, we established a reliable Mpro-NanoBiT assay for high-throughput screening inhibitors of SARS-CoV-2 main protease. Importantly, our methods can potentially assist in the rapid discovery of drug leads for the treatment of new emerging infectious diseases that currently lack specific drugs.
AVAILABILITY OF DATA AND MATERIALS
The data and materials developed in this article are available upon request.
FUNDING
This study was supported by Nantong Municipal Bureau of Science and Technology (MS12021038), Nantong University Talent Program (03083027).
REFERENCES
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265- 269.
- Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109: 102433.
- Liu K, Fang YY, Deng Y, Liu W, Wang MF, et al. (2020) Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 133(9): 1025-1031.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395: 497-506.
- Painter GR, Natchus MG, Cohen O, Holman W, Painter WP (2021) Developing a direct acting, orally available antiviral agent in a pandemic: The evolution of molnupiravir as a potential treatment for COVID-19. Curr Opin Virol 50: 17-22.
- Kabinger F, Stiller C, Schmitzova J, Dienemann C, Kokic G, et al. (2021) Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol 28: 740-746.
- Singh TU, Parida S, Lingaraju MC, Kesavan M, Kumar D, et al. (2020) Drug repurposing approach to fight COVID-19. Pharmacol Rep 72: 1479-1508.
- Zhang Y, Tang LV (2021) Overview of targets and potential drugs of SARS-CoV-2 according to the viral replication. J Proteome Res 20: 49-59.
- Ullrich S, Nitsche C (2020) The SARS-CoV-2 main protease as drug target. Bio org Med Chem Lett 30: 127377.
- Lin CW, Lin KH, Hsieh TH, Shiu SY, Li JY (2006) severe acute respiratory syndrome coronavirus 3C- like protease-induced apoptosis. FEMS Immunol Med Microbiol 46: 375-380.
- Rothan HA, Teoh TC (2021) Cell-based high-throughput screening protocol for discovering antiviral inhibitors against SARS-COV-2 main protease (3CLpro). Mol Biotechnol 63: 240-248.
- Mahmud S, Uddin MAR, Zaman M, Sujon KM, Rahman ME, et al. (2021) Molecular docking and dynamics study of natural compound for potential inhibition of main protease of SARS-CoV-2. J Biomol Struct Dyn 39: 6281-6289.
- Jin Z, Du X, Xu Y, Deng Y, Liu M, et al. (2020) Structure of Mpro from SARSCoV- 2 and discovery of its inhibitors. Nature 582: 289-293.
- Ong ILH, Yang KL (2017) Recent developments in protease activity assays and sensors. Analyst 142: 1867-1881.
- Drazic T, Kuhl N, Leuthold MM, Behnam MAM, Klein CD (2021) Efficiency improvements and discovery of new substrates for a SARS-CoV-2 main protease FRET assay. SLAS Discov 26: 1189-1199.
- Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, et al. (2014) MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42(Database issue): D297-303.
- Copeland RA (2003) Mechanistic considerations in high-throughput screening. Analytical Biochemistry 320: 1-12.
- Domkin V, Chabes A (2014) Phosphines are ribonucleotide reductase reductants that act via C- terminal cysteines similar to thioredoxins and glutaredoxins. Sci Rep 4: 5539.
- Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, et al. (2020) Boceprevir, GC- 376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 30: 678-692.
- Dai W, Zhang B, Jiang XM, Su H, Li J, et al. (2020) Structure- based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368: 1331-1335.
- Krishnaswamy S (2005) Exosite-driven substrate specificity and function in coagulation. J Thromb Haemost 3: 54-67.
- Green DR, Evan GI (2002) A matter of life and death. Cancer Cell 1: 19- 30.
- Hughes JP, Rees S, Kalindjian SB, Philpott KL (2011) Principles of early drug discovery. Br J Pharmacol 162: 1239-1249.
- Yang ZY, He JH, Lu AP, Hou TJ, Cao DS (2020) Frequent hitters: nuisance artifacts in high-throughput screening. Drug Discov Today 25: 657-667.
- Ghosh D, Koch U, Hadian K, Sattler M, Tetko IV (2018) Luciferase advisor: high-accuracy model to flag false positive hits in luciferase HTS assays. J Chem Inf Model 58: 933-942.
Article Type
Research Article
Publication history
Received Date: December 03, 2021
Published: December 10, 2021
Address for correspondence
Jianxi Liu, Institute of Special Environmental Medicine, Nantong University, China
Copyright
©2021 Open Access Journal of Biomedical Science, All rights reserved. No part of this content may be reproduced or transmitted in any form or by any means as per the standard guidelines of fair use. Open Access Journal of Biomedical Science is licensed under a Creative Commons Attribution 4.0 International License
How to cite this article
Zhiheng Z, Qingyu M, Xia L, Yide Z, Wei L, Jianxi L. Development of High Throughput Screening to Discover SARS-CoV-2 Main Protease Inhibitors with NanoBit Complementation. 2021- 3(6) OAJBS.ID.000360.
Figure 1: Design of Mpro-NanoBiT assay.
(A) Mpro cleavage site was fused into NanoBiT. The dark green part is the short peptide of SmBiT, while the light
green part is the long peptide of LgBiT, and the blue part is the Mpro cleavage site. When the Mpro cleavage site is
cleaved by Mpro, the subunits of two NanoLuc are separated, and enzyme activity is disabled.
(B) Detailed primary structure of Mpro cleavage site-NanoBiT. Yellow letters are amino acids of secretion signal
peptide. The dark green part is SmBiT, while the light green part is LgBiT, and the blue part is Mpro cleavage site.
Figure 2: Mpro cleavage site-NanoBiT was intact with NanoLuc activity and Mpro cleavage site was accessible to Mpro.
(A) Mpro cleavage site-NanoBiT was mainly secreted into cell culture medium.
(B) The molecular weight of the expressed Mpro cleavage site-NanoBiT was close to expected size, 34kD. Lane 1,
protein marker; Lane 2, Mpro cleavage site-NanoBiT.
(C) Mpro cleavage site in Mpro cleavage site-NanoBiT was able to be exposed to and digested by Mpro.
Figure 3: (A) Optimization of the dilution factor for Mpro cleavage site-NanoBiT. The purified protein was diluted in 20,
30, 40, 50, and 60 folds, respectively. Then the prepared luminescence substrate was added in a volume ratio of 1:1.
Signals were detected in 1, 5, 10 and 15min after substrate addition.
(B) Optimization of the concentration of SARS-CoV-2 Mpro. The appropriately diluted Mpro cleavage site-NanoBiT was
added into a 384-well plate, followed by addition of 0, 0.25, 0.5,1, 1.25μg of Mpro in a final volume to 25μL. Equal
volume of luminescence substrate was added, and the plates were read in 1, 5, 10, and 15min.
(C) Optimization of
DMSO concentration. Diluted Mpro cleavage site-NanoBiT plus 0, 0.25, 0.5, 1% DMSO was split into a 384-well plate,
followed by addition of 1μg of Mpro. The luminescence was measured in 1, 5, 10 and15 min.
(D) IC50s of GC376 and Mpro inhibitor 11a were measured in the reaction containing 60-fold diluted Mpro cleavage
site-NanoBiT, 1μg of Mpro, 1mM DTT and 0.5 % DMSO.
Figure 4: The processes of high throughput screening of SARS-CoV-2 Mpro inhibitors.
(1) Mpro was diluted at 1μg per 10μL with DMEM plus 2.5mM DTT and split into 384-well plates at 10μL/well with liquid
handler.
(2) 5μL of 100μM compounds were added into the plates, mixed and collected to the bottom of wells by
centrifuge. The plates were stored at room temperature for 30 min to allow compounds to fully bind to Mpro. A2-D2,
0.5% DMSO; M23-P23, 300nM GC376.
(3) 10μL of diluted Mpro cleavage site-NanoBiT was added, mixed and centrifuged.
(4) All plates were placed at 37 °C for 1.5 hours.
(5) 25μL of Nano luminescence substrate was added into each well, mixed and centrifuged.
(6) After incubation at room temperature for 10-20min, the plates were read. All signals were picked up with
integration time of 100 microseconds.
Figure 5: Evaluation of Mpro-NanoBiT assay.
(A) Each plate was set for 4 wells of positive (300nM GC376) and 4 wells of negative controls (0.5% DMSO). Twenty
plates of compounds were screened.
(B) 6,400 compounds were tested. Most were silent compounds which did not affect the function of Mpro and the
sample over control value are around 0.5. Compounds which have values more than 0.5 are considered having
inhibitory effect.